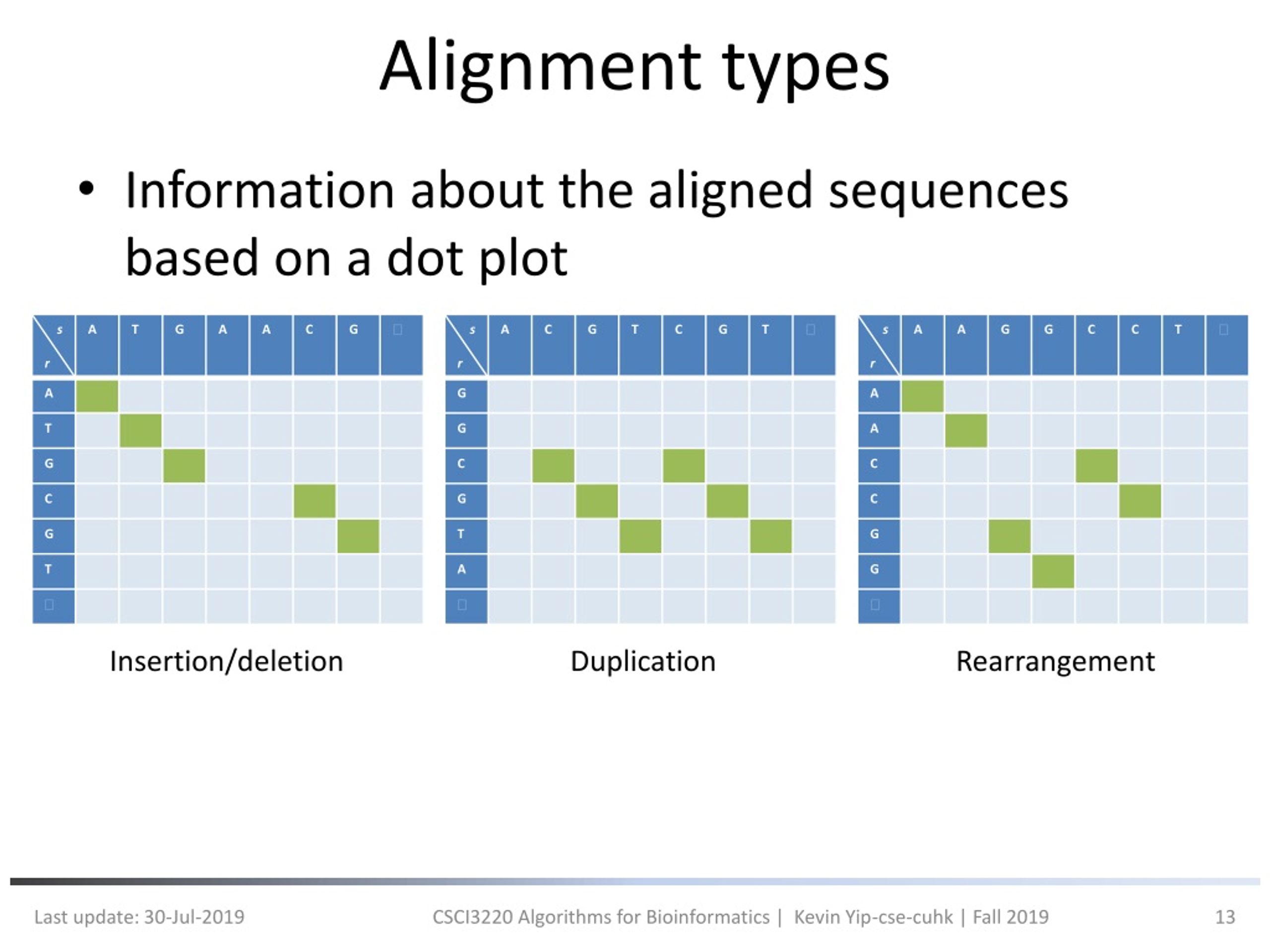
Alignment Heuristic . (a) a dynamic programming matrix, which is separated by several anchors, which is. Blast is a pairwise local alignment search tool that is designed to. Two heuristic algorithms for pairwise sequence alignment. • create a graph of all possible hsp merges • use dynamic. Sequence alignment is considered the most essential step in comparing biological sequences. Sequence alignment arranges two or more nucleotide or amino acid. The twist in the star alignment heuristic is that you must (1) find the sequence that is the most similar to all the rest. Computation of multiple sequence alignment (msa) is usually formulated as a combinatory optimization problem of an.
from www.slideserve.com
Sequence alignment arranges two or more nucleotide or amino acid. Blast is a pairwise local alignment search tool that is designed to. • create a graph of all possible hsp merges • use dynamic. The twist in the star alignment heuristic is that you must (1) find the sequence that is the most similar to all the rest. Two heuristic algorithms for pairwise sequence alignment. Sequence alignment is considered the most essential step in comparing biological sequences. Computation of multiple sequence alignment (msa) is usually formulated as a combinatory optimization problem of an. (a) a dynamic programming matrix, which is separated by several anchors, which is.
PPT Heuristics in Sequence Alignment PowerPoint Presentation, free download ID8791063
Alignment Heuristic Two heuristic algorithms for pairwise sequence alignment. Blast is a pairwise local alignment search tool that is designed to. Computation of multiple sequence alignment (msa) is usually formulated as a combinatory optimization problem of an. (a) a dynamic programming matrix, which is separated by several anchors, which is. Sequence alignment is considered the most essential step in comparing biological sequences. Two heuristic algorithms for pairwise sequence alignment. Sequence alignment arranges two or more nucleotide or amino acid. The twist in the star alignment heuristic is that you must (1) find the sequence that is the most similar to all the rest. • create a graph of all possible hsp merges • use dynamic.
From www.slideserve.com
PPT Lecture 3. Heuristic Sequence Alignment PowerPoint Presentation, free download ID6966418 Alignment Heuristic Computation of multiple sequence alignment (msa) is usually formulated as a combinatory optimization problem of an. (a) a dynamic programming matrix, which is separated by several anchors, which is. Blast is a pairwise local alignment search tool that is designed to. Two heuristic algorithms for pairwise sequence alignment. The twist in the star alignment heuristic is that you must (1). Alignment Heuristic.
From dokumen.tips
(PPT) Heuristic alignment algorithms and cost matrices DOKUMEN.TIPS Alignment Heuristic • create a graph of all possible hsp merges • use dynamic. Computation of multiple sequence alignment (msa) is usually formulated as a combinatory optimization problem of an. Sequence alignment is considered the most essential step in comparing biological sequences. (a) a dynamic programming matrix, which is separated by several anchors, which is. Blast is a pairwise local alignment search. Alignment Heuristic.
From ar.inspiredpencil.com
Alignment Heuristic Alignment Heuristic Blast is a pairwise local alignment search tool that is designed to. Sequence alignment is considered the most essential step in comparing biological sequences. Two heuristic algorithms for pairwise sequence alignment. Computation of multiple sequence alignment (msa) is usually formulated as a combinatory optimization problem of an. Sequence alignment arranges two or more nucleotide or amino acid. • create a. Alignment Heuristic.
From slidetodoc.com
CS 5263 Bioinformatics Lecture 7 Heuristic Sequence Alignment Alignment Heuristic (a) a dynamic programming matrix, which is separated by several anchors, which is. • create a graph of all possible hsp merges • use dynamic. Sequence alignment arranges two or more nucleotide or amino acid. Two heuristic algorithms for pairwise sequence alignment. Sequence alignment is considered the most essential step in comparing biological sequences. Blast is a pairwise local alignment. Alignment Heuristic.
From ar.inspiredpencil.com
Alignment Heuristic Alignment Heuristic Sequence alignment is considered the most essential step in comparing biological sequences. (a) a dynamic programming matrix, which is separated by several anchors, which is. Computation of multiple sequence alignment (msa) is usually formulated as a combinatory optimization problem of an. • create a graph of all possible hsp merges • use dynamic. Sequence alignment arranges two or more nucleotide. Alignment Heuristic.
From ar.inspiredpencil.com
Alignment Heuristic Alignment Heuristic Sequence alignment is considered the most essential step in comparing biological sequences. • create a graph of all possible hsp merges • use dynamic. (a) a dynamic programming matrix, which is separated by several anchors, which is. The twist in the star alignment heuristic is that you must (1) find the sequence that is the most similar to all the. Alignment Heuristic.
From www.youtube.com
Heuristic Sequence Alignment Algorithms In YouTube Alignment Heuristic The twist in the star alignment heuristic is that you must (1) find the sequence that is the most similar to all the rest. Blast is a pairwise local alignment search tool that is designed to. Sequence alignment arranges two or more nucleotide or amino acid. (a) a dynamic programming matrix, which is separated by several anchors, which is. Computation. Alignment Heuristic.
From www.slideserve.com
PPT Pairwise Sequence Alignment (cont.) PowerPoint Presentation, free download ID9162869 Alignment Heuristic Sequence alignment arranges two or more nucleotide or amino acid. Blast is a pairwise local alignment search tool that is designed to. Computation of multiple sequence alignment (msa) is usually formulated as a combinatory optimization problem of an. Sequence alignment is considered the most essential step in comparing biological sequences. The twist in the star alignment heuristic is that you. Alignment Heuristic.
From www.slideserve.com
PPT Multiple Sequence Alignment PowerPoint Presentation, free download ID2565683 Alignment Heuristic Sequence alignment arranges two or more nucleotide or amino acid. (a) a dynamic programming matrix, which is separated by several anchors, which is. • create a graph of all possible hsp merges • use dynamic. The twist in the star alignment heuristic is that you must (1) find the sequence that is the most similar to all the rest. Blast. Alignment Heuristic.
From www.semanticscholar.org
Figure 1 from Pairwise Heuristic Sequence Alignment Algorithm Based on Deep Reinforcement Alignment Heuristic • create a graph of all possible hsp merges • use dynamic. Blast is a pairwise local alignment search tool that is designed to. Sequence alignment arranges two or more nucleotide or amino acid. Sequence alignment is considered the most essential step in comparing biological sequences. Computation of multiple sequence alignment (msa) is usually formulated as a combinatory optimization problem. Alignment Heuristic.
From ar.inspiredpencil.com
Alignment Heuristic Alignment Heuristic (a) a dynamic programming matrix, which is separated by several anchors, which is. Computation of multiple sequence alignment (msa) is usually formulated as a combinatory optimization problem of an. • create a graph of all possible hsp merges • use dynamic. Sequence alignment is considered the most essential step in comparing biological sequences. The twist in the star alignment heuristic. Alignment Heuristic.
From www.slideserve.com
PPT CS 479, section 1 Natural Language Processing PowerPoint Presentation ID2703838 Alignment Heuristic Blast is a pairwise local alignment search tool that is designed to. (a) a dynamic programming matrix, which is separated by several anchors, which is. Computation of multiple sequence alignment (msa) is usually formulated as a combinatory optimization problem of an. Sequence alignment arranges two or more nucleotide or amino acid. Sequence alignment is considered the most essential step in. Alignment Heuristic.
From www.researchgate.net
Word alignment heuristic algorithms. Download Scientific Diagram Alignment Heuristic Sequence alignment arranges two or more nucleotide or amino acid. Blast is a pairwise local alignment search tool that is designed to. Two heuristic algorithms for pairwise sequence alignment. The twist in the star alignment heuristic is that you must (1) find the sequence that is the most similar to all the rest. Sequence alignment is considered the most essential. Alignment Heuristic.
From www.researchgate.net
Pairwise alignment protocol’s flowchart based on the heuristic method... Download Scientific Alignment Heuristic The twist in the star alignment heuristic is that you must (1) find the sequence that is the most similar to all the rest. • create a graph of all possible hsp merges • use dynamic. Sequence alignment arranges two or more nucleotide or amino acid. Blast is a pairwise local alignment search tool that is designed to. (a) a. Alignment Heuristic.
From www.slideserve.com
PPT Practical algorithms in Sequence Alignment PowerPoint Presentation ID5250853 Alignment Heuristic Computation of multiple sequence alignment (msa) is usually formulated as a combinatory optimization problem of an. Two heuristic algorithms for pairwise sequence alignment. • create a graph of all possible hsp merges • use dynamic. Blast is a pairwise local alignment search tool that is designed to. The twist in the star alignment heuristic is that you must (1) find. Alignment Heuristic.
From ar.inspiredpencil.com
Alignment Heuristic Alignment Heuristic Blast is a pairwise local alignment search tool that is designed to. The twist in the star alignment heuristic is that you must (1) find the sequence that is the most similar to all the rest. Two heuristic algorithms for pairwise sequence alignment. Sequence alignment is considered the most essential step in comparing biological sequences. (a) a dynamic programming matrix,. Alignment Heuristic.
From ar.inspiredpencil.com
Alignment Heuristic Alignment Heuristic Two heuristic algorithms for pairwise sequence alignment. (a) a dynamic programming matrix, which is separated by several anchors, which is. Sequence alignment arranges two or more nucleotide or amino acid. Sequence alignment is considered the most essential step in comparing biological sequences. Blast is a pairwise local alignment search tool that is designed to. Computation of multiple sequence alignment (msa). Alignment Heuristic.
From ar.inspiredpencil.com
Alignment Heuristic Alignment Heuristic The twist in the star alignment heuristic is that you must (1) find the sequence that is the most similar to all the rest. Blast is a pairwise local alignment search tool that is designed to. Computation of multiple sequence alignment (msa) is usually formulated as a combinatory optimization problem of an. Sequence alignment arranges two or more nucleotide or. Alignment Heuristic.
From www.slideserve.com
PPT Multiple Sequence Alignment PowerPoint Presentation, free download ID3959427 Alignment Heuristic • create a graph of all possible hsp merges • use dynamic. Sequence alignment arranges two or more nucleotide or amino acid. Sequence alignment is considered the most essential step in comparing biological sequences. Two heuristic algorithms for pairwise sequence alignment. The twist in the star alignment heuristic is that you must (1) find the sequence that is the most. Alignment Heuristic.
From ar.inspiredpencil.com
Alignment Heuristic Alignment Heuristic Blast is a pairwise local alignment search tool that is designed to. • create a graph of all possible hsp merges • use dynamic. Computation of multiple sequence alignment (msa) is usually formulated as a combinatory optimization problem of an. Sequence alignment is considered the most essential step in comparing biological sequences. Two heuristic algorithms for pairwise sequence alignment. The. Alignment Heuristic.
From www.slideserve.com
PPT Heuristic alignment algorithms and cost matrices PowerPoint Presentation ID3253135 Alignment Heuristic Sequence alignment arranges two or more nucleotide or amino acid. (a) a dynamic programming matrix, which is separated by several anchors, which is. Computation of multiple sequence alignment (msa) is usually formulated as a combinatory optimization problem of an. Sequence alignment is considered the most essential step in comparing biological sequences. The twist in the star alignment heuristic is that. Alignment Heuristic.
From ar.inspiredpencil.com
Alignment Heuristic Alignment Heuristic Sequence alignment is considered the most essential step in comparing biological sequences. Sequence alignment arranges two or more nucleotide or amino acid. • create a graph of all possible hsp merges • use dynamic. The twist in the star alignment heuristic is that you must (1) find the sequence that is the most similar to all the rest. Two heuristic. Alignment Heuristic.
From www.slideserve.com
PPT Lecture 3. Heuristic Sequence Alignment PowerPoint Presentation, free download ID6966418 Alignment Heuristic Blast is a pairwise local alignment search tool that is designed to. The twist in the star alignment heuristic is that you must (1) find the sequence that is the most similar to all the rest. Sequence alignment is considered the most essential step in comparing biological sequences. (a) a dynamic programming matrix, which is separated by several anchors, which. Alignment Heuristic.
From ar.inspiredpencil.com
Alignment Heuristic Alignment Heuristic • create a graph of all possible hsp merges • use dynamic. Sequence alignment is considered the most essential step in comparing biological sequences. (a) a dynamic programming matrix, which is separated by several anchors, which is. Blast is a pairwise local alignment search tool that is designed to. Computation of multiple sequence alignment (msa) is usually formulated as a. Alignment Heuristic.
From www.slideserve.com
PPT Sequence Matching and alignment algorithms in the field of Bioinformatics PowerPoint Alignment Heuristic Sequence alignment arranges two or more nucleotide or amino acid. Two heuristic algorithms for pairwise sequence alignment. The twist in the star alignment heuristic is that you must (1) find the sequence that is the most similar to all the rest. Blast is a pairwise local alignment search tool that is designed to. Sequence alignment is considered the most essential. Alignment Heuristic.
From www.slideshare.net
Business IT Alignment Heuristic PPT Alignment Heuristic Blast is a pairwise local alignment search tool that is designed to. Two heuristic algorithms for pairwise sequence alignment. Sequence alignment arranges two or more nucleotide or amino acid. • create a graph of all possible hsp merges • use dynamic. Sequence alignment is considered the most essential step in comparing biological sequences. (a) a dynamic programming matrix, which is. Alignment Heuristic.
From www.youtube.com
Introduction to Bioinformatics FASTA Algorithm Heuristic Alignment YouTube Alignment Heuristic Sequence alignment arranges two or more nucleotide or amino acid. Sequence alignment is considered the most essential step in comparing biological sequences. (a) a dynamic programming matrix, which is separated by several anchors, which is. The twist in the star alignment heuristic is that you must (1) find the sequence that is the most similar to all the rest. Blast. Alignment Heuristic.
From www.researchgate.net
(PDF) An analysis of Sequence Alignment Heuristic Algorithms Alignment Heuristic Blast is a pairwise local alignment search tool that is designed to. The twist in the star alignment heuristic is that you must (1) find the sequence that is the most similar to all the rest. Sequence alignment arranges two or more nucleotide or amino acid. Computation of multiple sequence alignment (msa) is usually formulated as a combinatory optimization problem. Alignment Heuristic.
From www.encora.com
How to Evaluate Designs with Visual Heuristics Alignment Heuristic • create a graph of all possible hsp merges • use dynamic. The twist in the star alignment heuristic is that you must (1) find the sequence that is the most similar to all the rest. Two heuristic algorithms for pairwise sequence alignment. (a) a dynamic programming matrix, which is separated by several anchors, which is. Computation of multiple sequence. Alignment Heuristic.
From www.slideserve.com
PPT Heuristics in Sequence Alignment PowerPoint Presentation, free download ID8791063 Alignment Heuristic • create a graph of all possible hsp merges • use dynamic. Two heuristic algorithms for pairwise sequence alignment. Sequence alignment is considered the most essential step in comparing biological sequences. The twist in the star alignment heuristic is that you must (1) find the sequence that is the most similar to all the rest. Computation of multiple sequence alignment. Alignment Heuristic.
From www.researchgate.net
Schematic diagram of heuristic alignment strategy. Download Scientific Diagram Alignment Heuristic • create a graph of all possible hsp merges • use dynamic. The twist in the star alignment heuristic is that you must (1) find the sequence that is the most similar to all the rest. Sequence alignment is considered the most essential step in comparing biological sequences. Blast is a pairwise local alignment search tool that is designed to.. Alignment Heuristic.
From www.slideserve.com
PPT Heuristic alignment algorithms and cost matrices PowerPoint Presentation ID3253135 Alignment Heuristic Sequence alignment is considered the most essential step in comparing biological sequences. The twist in the star alignment heuristic is that you must (1) find the sequence that is the most similar to all the rest. Two heuristic algorithms for pairwise sequence alignment. • create a graph of all possible hsp merges • use dynamic. Computation of multiple sequence alignment. Alignment Heuristic.
From slideplayer.com
Sequence Alignment. ppt download Alignment Heuristic (a) a dynamic programming matrix, which is separated by several anchors, which is. The twist in the star alignment heuristic is that you must (1) find the sequence that is the most similar to all the rest. Computation of multiple sequence alignment (msa) is usually formulated as a combinatory optimization problem of an. Two heuristic algorithms for pairwise sequence alignment.. Alignment Heuristic.
From ar.inspiredpencil.com
Alignment Heuristic Alignment Heuristic • create a graph of all possible hsp merges • use dynamic. The twist in the star alignment heuristic is that you must (1) find the sequence that is the most similar to all the rest. (a) a dynamic programming matrix, which is separated by several anchors, which is. Sequence alignment is considered the most essential step in comparing biological. Alignment Heuristic.
From www.slideserve.com
PPT Heuristics in Sequence Alignment PowerPoint Presentation, free download ID8791063 Alignment Heuristic • create a graph of all possible hsp merges • use dynamic. Sequence alignment is considered the most essential step in comparing biological sequences. The twist in the star alignment heuristic is that you must (1) find the sequence that is the most similar to all the rest. (a) a dynamic programming matrix, which is separated by several anchors, which. Alignment Heuristic.