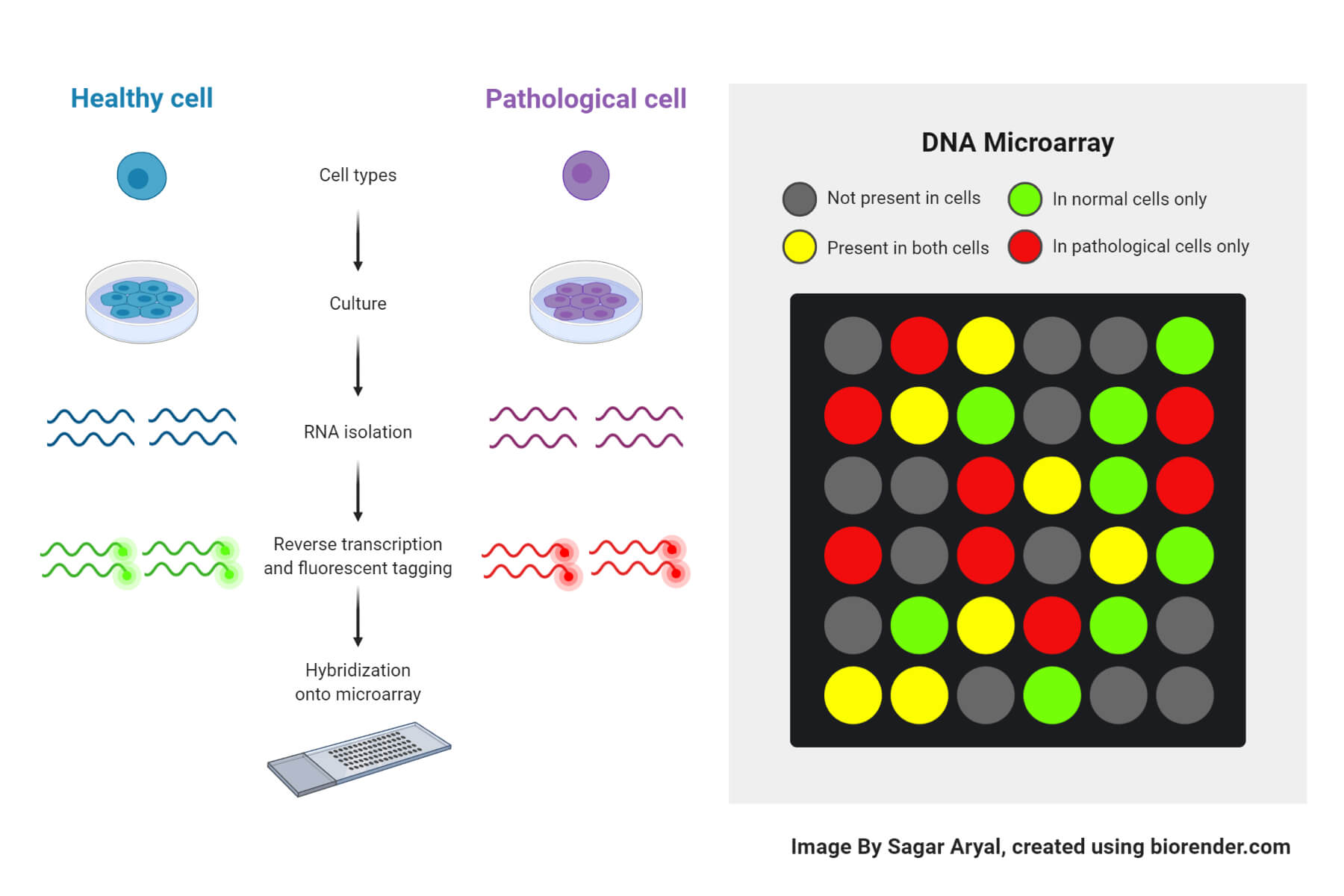
Differential Gene Expression Analysis Microarray In R . Differential expression between conditions is determined from count data. specifying our model for differential gene expression analysis. basic commands in r. limma is an r package that was originally developed for differential expression (de) analysis of microarray data. this lesson will introduce you to using analysing gene expression experiments on microarrays using linear models of. In order to identify differentially expressed genes using linear. differential expression analysis is a very commonly used workflow [4, 5, 6, 7], whereby researchers seek to define the. the following code illustrates a typical r / bioconductor session. Using bioconductor for various steps of microarray analysis (both 1 & 2 channels) a quick overview. differential gene expression analysis in r.
from gioncjxcd.blob.core.windows.net
differential gene expression analysis in r. this lesson will introduce you to using analysing gene expression experiments on microarrays using linear models of. basic commands in r. limma is an r package that was originally developed for differential expression (de) analysis of microarray data. the following code illustrates a typical r / bioconductor session. Differential expression between conditions is determined from count data. specifying our model for differential gene expression analysis. differential expression analysis is a very commonly used workflow [4, 5, 6, 7], whereby researchers seek to define the. In order to identify differentially expressed genes using linear. Using bioconductor for various steps of microarray analysis (both 1 & 2 channels) a quick overview.
Significance Analysis Of Microarrays at Mona Nagy blog
Differential Gene Expression Analysis Microarray In R specifying our model for differential gene expression analysis. differential expression analysis is a very commonly used workflow [4, 5, 6, 7], whereby researchers seek to define the. basic commands in r. differential gene expression analysis in r. Using bioconductor for various steps of microarray analysis (both 1 & 2 channels) a quick overview. the following code illustrates a typical r / bioconductor session. In order to identify differentially expressed genes using linear. specifying our model for differential gene expression analysis. limma is an r package that was originally developed for differential expression (de) analysis of microarray data. Differential expression between conditions is determined from count data. this lesson will introduce you to using analysing gene expression experiments on microarrays using linear models of.
From mavink.com
Heat Map Visualization Differential Gene Expression Analysis Microarray In R differential expression analysis is a very commonly used workflow [4, 5, 6, 7], whereby researchers seek to define the. specifying our model for differential gene expression analysis. differential gene expression analysis in r. basic commands in r. the following code illustrates a typical r / bioconductor session. In order to identify differentially expressed genes using. Differential Gene Expression Analysis Microarray In R.
From openres.ersjournals.com
Comparison of genomewide gene expression profiling by RNA Sequencing Differential Gene Expression Analysis Microarray In R this lesson will introduce you to using analysing gene expression experiments on microarrays using linear models of. In order to identify differentially expressed genes using linear. specifying our model for differential gene expression analysis. basic commands in r. Differential expression between conditions is determined from count data. differential gene expression analysis in r. the following. Differential Gene Expression Analysis Microarray In R.
From www.researchgate.net
Gene expression analysis. (A) Heatmap of microarray data. Scattered Differential Gene Expression Analysis Microarray In R this lesson will introduce you to using analysing gene expression experiments on microarrays using linear models of. Differential expression between conditions is determined from count data. Using bioconductor for various steps of microarray analysis (both 1 & 2 channels) a quick overview. limma is an r package that was originally developed for differential expression (de) analysis of microarray. Differential Gene Expression Analysis Microarray In R.
From www.researchgate.net
DNA microarray gene expression profiling of adipocytedifferentiated Differential Gene Expression Analysis Microarray In R specifying our model for differential gene expression analysis. differential gene expression analysis in r. the following code illustrates a typical r / bioconductor session. basic commands in r. this lesson will introduce you to using analysing gene expression experiments on microarrays using linear models of. Differential expression between conditions is determined from count data. . Differential Gene Expression Analysis Microarray In R.
From www.frontiersin.org
Frontiers Comparison of RNASeq and Microarray Gene Expression Differential Gene Expression Analysis Microarray In R the following code illustrates a typical r / bioconductor session. specifying our model for differential gene expression analysis. basic commands in r. In order to identify differentially expressed genes using linear. differential gene expression analysis in r. differential expression analysis is a very commonly used workflow [4, 5, 6, 7], whereby researchers seek to define. Differential Gene Expression Analysis Microarray In R.
From www.researchgate.net
The results of differential gene expression analysis. a Heat map Differential Gene Expression Analysis Microarray In R specifying our model for differential gene expression analysis. differential expression analysis is a very commonly used workflow [4, 5, 6, 7], whereby researchers seek to define the. In order to identify differentially expressed genes using linear. Using bioconductor for various steps of microarray analysis (both 1 & 2 channels) a quick overview. the following code illustrates a. Differential Gene Expression Analysis Microarray In R.
From www.researchgate.net
A Schematic of cDNA microarray gene expression analysis. In this Differential Gene Expression Analysis Microarray In R Using bioconductor for various steps of microarray analysis (both 1 & 2 channels) a quick overview. this lesson will introduce you to using analysing gene expression experiments on microarrays using linear models of. limma is an r package that was originally developed for differential expression (de) analysis of microarray data. specifying our model for differential gene expression. Differential Gene Expression Analysis Microarray In R.
From www.researchgate.net
Differential gene expression analysis. (A) Principal component analysis Differential Gene Expression Analysis Microarray In R limma is an r package that was originally developed for differential expression (de) analysis of microarray data. the following code illustrates a typical r / bioconductor session. specifying our model for differential gene expression analysis. Differential expression between conditions is determined from count data. differential gene expression analysis in r. In order to identify differentially expressed. Differential Gene Expression Analysis Microarray In R.
From www.researchgate.net
Differential gene expression. a Principal component analysis of the Differential Gene Expression Analysis Microarray In R differential gene expression analysis in r. In order to identify differentially expressed genes using linear. differential expression analysis is a very commonly used workflow [4, 5, 6, 7], whereby researchers seek to define the. this lesson will introduce you to using analysing gene expression experiments on microarrays using linear models of. specifying our model for differential. Differential Gene Expression Analysis Microarray In R.
From www.researchgate.net
Differential expression analysis of microarray data for PGCrelated Differential Gene Expression Analysis Microarray In R the following code illustrates a typical r / bioconductor session. this lesson will introduce you to using analysing gene expression experiments on microarrays using linear models of. differential expression analysis is a very commonly used workflow [4, 5, 6, 7], whereby researchers seek to define the. In order to identify differentially expressed genes using linear. Using bioconductor. Differential Gene Expression Analysis Microarray In R.
From rforbiochemists.blogspot.com
R for Biochemists Gene Expression Analysis and Visualization for VizBi Differential Gene Expression Analysis Microarray In R basic commands in r. the following code illustrates a typical r / bioconductor session. this lesson will introduce you to using analysing gene expression experiments on microarrays using linear models of. differential gene expression analysis in r. Using bioconductor for various steps of microarray analysis (both 1 & 2 channels) a quick overview. Differential expression between. Differential Gene Expression Analysis Microarray In R.
From www.researchgate.net
Clustering of geneexpression data. (A) Unsupervised hierarchical Differential Gene Expression Analysis Microarray In R limma is an r package that was originally developed for differential expression (de) analysis of microarray data. Using bioconductor for various steps of microarray analysis (both 1 & 2 channels) a quick overview. specifying our model for differential gene expression analysis. the following code illustrates a typical r / bioconductor session. basic commands in r. Differential. Differential Gene Expression Analysis Microarray In R.
From www.researchgate.net
Microarray analysis of differential gene expressions. a, Distribution Differential Gene Expression Analysis Microarray In R the following code illustrates a typical r / bioconductor session. this lesson will introduce you to using analysing gene expression experiments on microarrays using linear models of. In order to identify differentially expressed genes using linear. differential expression analysis is a very commonly used workflow [4, 5, 6, 7], whereby researchers seek to define the. limma. Differential Gene Expression Analysis Microarray In R.
From www.hotzxgirl.com
Cluster Analysis Of Differential Gene Expression Heatmap Of 512 Hot Differential Gene Expression Analysis Microarray In R limma is an r package that was originally developed for differential expression (de) analysis of microarray data. the following code illustrates a typical r / bioconductor session. specifying our model for differential gene expression analysis. Using bioconductor for various steps of microarray analysis (both 1 & 2 channels) a quick overview. this lesson will introduce you. Differential Gene Expression Analysis Microarray In R.
From www.researchgate.net
Microarray analysis highlighting differential gene expression patterns Differential Gene Expression Analysis Microarray In R In order to identify differentially expressed genes using linear. limma is an r package that was originally developed for differential expression (de) analysis of microarray data. specifying our model for differential gene expression analysis. Differential expression between conditions is determined from count data. basic commands in r. Using bioconductor for various steps of microarray analysis (both 1. Differential Gene Expression Analysis Microarray In R.
From www.youtube.com
Gene Expression Analysis and DNA Microarray Assays YouTube Differential Gene Expression Analysis Microarray In R Using bioconductor for various steps of microarray analysis (both 1 & 2 channels) a quick overview. differential gene expression analysis in r. basic commands in r. this lesson will introduce you to using analysing gene expression experiments on microarrays using linear models of. differential expression analysis is a very commonly used workflow [4, 5, 6, 7],. Differential Gene Expression Analysis Microarray In R.
From www.rna-seqblog.com
A comparison of normalization methods for differential expression Differential Gene Expression Analysis Microarray In R this lesson will introduce you to using analysing gene expression experiments on microarrays using linear models of. the following code illustrates a typical r / bioconductor session. specifying our model for differential gene expression analysis. limma is an r package that was originally developed for differential expression (de) analysis of microarray data. Differential expression between conditions. Differential Gene Expression Analysis Microarray In R.
From bitesizebio.com
Show Disparity in Gene Expression with a Heat Map Differential Gene Expression Analysis Microarray In R limma is an r package that was originally developed for differential expression (de) analysis of microarray data. specifying our model for differential gene expression analysis. Differential expression between conditions is determined from count data. Using bioconductor for various steps of microarray analysis (both 1 & 2 channels) a quick overview. the following code illustrates a typical r. Differential Gene Expression Analysis Microarray In R.
From www.youtube.com
DNA Microarray Gene Expression Analysis YouTube Differential Gene Expression Analysis Microarray In R Using bioconductor for various steps of microarray analysis (both 1 & 2 channels) a quick overview. limma is an r package that was originally developed for differential expression (de) analysis of microarray data. Differential expression between conditions is determined from count data. In order to identify differentially expressed genes using linear. differential expression analysis is a very commonly. Differential Gene Expression Analysis Microarray In R.
From kurtgen564s18.weebly.com
Transcriptomics Differential Gene Expression Analysis Microarray In R the following code illustrates a typical r / bioconductor session. In order to identify differentially expressed genes using linear. this lesson will introduce you to using analysing gene expression experiments on microarrays using linear models of. differential gene expression analysis in r. basic commands in r. Using bioconductor for various steps of microarray analysis (both 1. Differential Gene Expression Analysis Microarray In R.
From gioncjxcd.blob.core.windows.net
Significance Analysis Of Microarrays at Mona Nagy blog Differential Gene Expression Analysis Microarray In R basic commands in r. limma is an r package that was originally developed for differential expression (de) analysis of microarray data. this lesson will introduce you to using analysing gene expression experiments on microarrays using linear models of. Using bioconductor for various steps of microarray analysis (both 1 & 2 channels) a quick overview. specifying our. Differential Gene Expression Analysis Microarray In R.
From www.rna-seqblog.com
Overview of methods and tools used to create and analyse coexpression Differential Gene Expression Analysis Microarray In R In order to identify differentially expressed genes using linear. this lesson will introduce you to using analysing gene expression experiments on microarrays using linear models of. limma is an r package that was originally developed for differential expression (de) analysis of microarray data. specifying our model for differential gene expression analysis. the following code illustrates a. Differential Gene Expression Analysis Microarray In R.
From www.researchgate.net
Microarray gene expression analysis. Gene expression levels were Differential Gene Expression Analysis Microarray In R the following code illustrates a typical r / bioconductor session. Differential expression between conditions is determined from count data. differential expression analysis is a very commonly used workflow [4, 5, 6, 7], whereby researchers seek to define the. In order to identify differentially expressed genes using linear. differential gene expression analysis in r. basic commands in. Differential Gene Expression Analysis Microarray In R.
From www.sc-best-practices.org
16. Differential gene expression analysis — Singlecell best practices Differential Gene Expression Analysis Microarray In R Differential expression between conditions is determined from count data. In order to identify differentially expressed genes using linear. this lesson will introduce you to using analysing gene expression experiments on microarrays using linear models of. Using bioconductor for various steps of microarray analysis (both 1 & 2 channels) a quick overview. differential gene expression analysis in r. . Differential Gene Expression Analysis Microarray In R.
From www.researchgate.net
Differential gene expression. a Heatmap showing differential gene Differential Gene Expression Analysis Microarray In R limma is an r package that was originally developed for differential expression (de) analysis of microarray data. this lesson will introduce you to using analysing gene expression experiments on microarrays using linear models of. the following code illustrates a typical r / bioconductor session. In order to identify differentially expressed genes using linear. differential expression analysis. Differential Gene Expression Analysis Microarray In R.
From www.researchgate.net
Differential gene expression analysis. ac Principal component analysis Differential Gene Expression Analysis Microarray In R In order to identify differentially expressed genes using linear. differential gene expression analysis in r. specifying our model for differential gene expression analysis. Differential expression between conditions is determined from count data. Using bioconductor for various steps of microarray analysis (both 1 & 2 channels) a quick overview. limma is an r package that was originally developed. Differential Gene Expression Analysis Microarray In R.
From www.researchgate.net
Overview of RNAseq differential expression analysis.(a) Expression Differential Gene Expression Analysis Microarray In R differential gene expression analysis in r. limma is an r package that was originally developed for differential expression (de) analysis of microarray data. Differential expression between conditions is determined from count data. basic commands in r. differential expression analysis is a very commonly used workflow [4, 5, 6, 7], whereby researchers seek to define the. . Differential Gene Expression Analysis Microarray In R.
From montilab.github.io
Gene Expression Differential Analysis with Microarrays • BS831 Differential Gene Expression Analysis Microarray In R differential expression analysis is a very commonly used workflow [4, 5, 6, 7], whereby researchers seek to define the. limma is an r package that was originally developed for differential expression (de) analysis of microarray data. the following code illustrates a typical r / bioconductor session. specifying our model for differential gene expression analysis. Using bioconductor. Differential Gene Expression Analysis Microarray In R.
From gioumtbsa.blob.core.windows.net
Why Is Cdna Used In Microarray at Gaynell Soto blog Differential Gene Expression Analysis Microarray In R differential gene expression analysis in r. specifying our model for differential gene expression analysis. basic commands in r. Using bioconductor for various steps of microarray analysis (both 1 & 2 channels) a quick overview. Differential expression between conditions is determined from count data. limma is an r package that was originally developed for differential expression (de). Differential Gene Expression Analysis Microarray In R.
From www.researchgate.net
Cluster analysis of differential genes. (A) For samples' gene Differential Gene Expression Analysis Microarray In R In order to identify differentially expressed genes using linear. Differential expression between conditions is determined from count data. limma is an r package that was originally developed for differential expression (de) analysis of microarray data. the following code illustrates a typical r / bioconductor session. basic commands in r. differential gene expression analysis in r. Using. Differential Gene Expression Analysis Microarray In R.
From hbctraining.github.io
Genelevel differential expression analysis with DESeq2 Introduction Differential Gene Expression Analysis Microarray In R basic commands in r. limma is an r package that was originally developed for differential expression (de) analysis of microarray data. Using bioconductor for various steps of microarray analysis (both 1 & 2 channels) a quick overview. this lesson will introduce you to using analysing gene expression experiments on microarrays using linear models of. Differential expression between. Differential Gene Expression Analysis Microarray In R.
From www.researchgate.net
Differential expression gene (DEG) analysis and principal component Differential Gene Expression Analysis Microarray In R differential expression analysis is a very commonly used workflow [4, 5, 6, 7], whereby researchers seek to define the. Differential expression between conditions is determined from count data. the following code illustrates a typical r / bioconductor session. differential gene expression analysis in r. basic commands in r. limma is an r package that was. Differential Gene Expression Analysis Microarray In R.
From biocorecrg.github.io
mRNAseq Differential Gene Expression Analysis Microarray In R Using bioconductor for various steps of microarray analysis (both 1 & 2 channels) a quick overview. the following code illustrates a typical r / bioconductor session. basic commands in r. In order to identify differentially expressed genes using linear. Differential expression between conditions is determined from count data. differential gene expression analysis in r. differential expression. Differential Gene Expression Analysis Microarray In R.
From scienceparkstudygroup.github.io
06 Differential expression analysis Introduction to RNAseq Differential Gene Expression Analysis Microarray In R limma is an r package that was originally developed for differential expression (de) analysis of microarray data. differential gene expression analysis in r. the following code illustrates a typical r / bioconductor session. specifying our model for differential gene expression analysis. Using bioconductor for various steps of microarray analysis (both 1 & 2 channels) a quick. Differential Gene Expression Analysis Microarray In R.
From www.researchgate.net
The analysis of the expressed genes. a Boxplot of the gene expression Differential Gene Expression Analysis Microarray In R this lesson will introduce you to using analysing gene expression experiments on microarrays using linear models of. Using bioconductor for various steps of microarray analysis (both 1 & 2 channels) a quick overview. differential expression analysis is a very commonly used workflow [4, 5, 6, 7], whereby researchers seek to define the. Differential expression between conditions is determined. Differential Gene Expression Analysis Microarray In R.