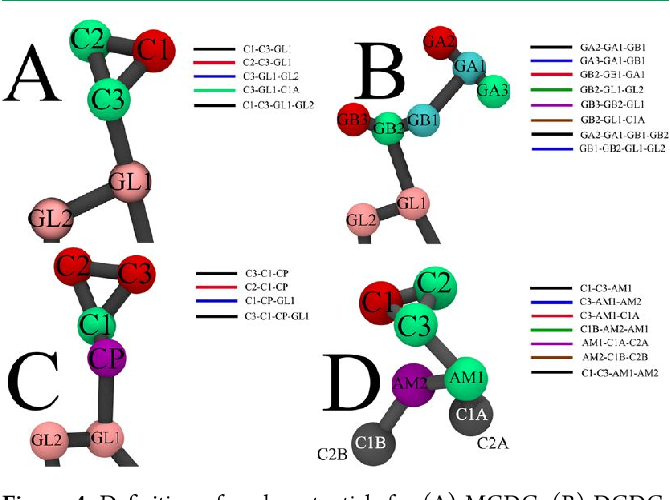
Martini Force Field Parameters For Glycolipids . bonded parameters are optimized by comparison to lipid conformations sampled with an atomistic force field, in particular with respect to the representation of the most populated states around the glycosidic linkage. based on this mapping, bonded parameters were derived by the standard procedure of fitting cg probability distributions of the. César lópez, zofie sovova, floris j. martini force field parameters to glycolipids. parametrization of the glycolipid force field, which is therefore fully compatible with the martini lipid,38 protein,42 and. bond and angle parameter for the new qa bead was adopted from the parameter of phosphatidylinositol (pi) in.
from www.semanticscholar.org
based on this mapping, bonded parameters were derived by the standard procedure of fitting cg probability distributions of the. bond and angle parameter for the new qa bead was adopted from the parameter of phosphatidylinositol (pi) in. parametrization of the glycolipid force field, which is therefore fully compatible with the martini lipid,38 protein,42 and. martini force field parameters to glycolipids. César lópez, zofie sovova, floris j. bonded parameters are optimized by comparison to lipid conformations sampled with an atomistic force field, in particular with respect to the representation of the most populated states around the glycosidic linkage.
Figure 4 from Martini Force Field Parameters for Glycolipids
Martini Force Field Parameters For Glycolipids parametrization of the glycolipid force field, which is therefore fully compatible with the martini lipid,38 protein,42 and. parametrization of the glycolipid force field, which is therefore fully compatible with the martini lipid,38 protein,42 and. César lópez, zofie sovova, floris j. bonded parameters are optimized by comparison to lipid conformations sampled with an atomistic force field, in particular with respect to the representation of the most populated states around the glycosidic linkage. bond and angle parameter for the new qa bead was adopted from the parameter of phosphatidylinositol (pi) in. martini force field parameters to glycolipids. based on this mapping, bonded parameters were derived by the standard procedure of fitting cg probability distributions of the.
From pubs.acs.org
Automatic Optimization of Lipid Models in the Martini Force Field Using Martini Force Field Parameters For Glycolipids bond and angle parameter for the new qa bead was adopted from the parameter of phosphatidylinositol (pi) in. bonded parameters are optimized by comparison to lipid conformations sampled with an atomistic force field, in particular with respect to the representation of the most populated states around the glycosidic linkage. martini force field parameters to glycolipids. based. Martini Force Field Parameters For Glycolipids.
From www.researchgate.net
Parameters of the DummyAssisted Dihedral for the MARTINI Protein Force Martini Force Field Parameters For Glycolipids parametrization of the glycolipid force field, which is therefore fully compatible with the martini lipid,38 protein,42 and. bond and angle parameter for the new qa bead was adopted from the parameter of phosphatidylinositol (pi) in. César lópez, zofie sovova, floris j. based on this mapping, bonded parameters were derived by the standard procedure of fitting cg probability. Martini Force Field Parameters For Glycolipids.
From www.researchgate.net
Topologies of asphaltene molecules and their Martini force field Martini Force Field Parameters For Glycolipids César lópez, zofie sovova, floris j. parametrization of the glycolipid force field, which is therefore fully compatible with the martini lipid,38 protein,42 and. martini force field parameters to glycolipids. bond and angle parameter for the new qa bead was adopted from the parameter of phosphatidylinositol (pi) in. bonded parameters are optimized by comparison to lipid conformations. Martini Force Field Parameters For Glycolipids.
From pubs.acs.org
Martini 3 Force Field Parameters for Protein Lipidation Post Martini Force Field Parameters For Glycolipids bond and angle parameter for the new qa bead was adopted from the parameter of phosphatidylinositol (pi) in. based on this mapping, bonded parameters were derived by the standard procedure of fitting cg probability distributions of the. martini force field parameters to glycolipids. César lópez, zofie sovova, floris j. parametrization of the glycolipid force field, which. Martini Force Field Parameters For Glycolipids.
From www.researchgate.net
Based on the MARTINI force field coarse granulation rules; (a Martini Force Field Parameters For Glycolipids parametrization of the glycolipid force field, which is therefore fully compatible with the martini lipid,38 protein,42 and. bonded parameters are optimized by comparison to lipid conformations sampled with an atomistic force field, in particular with respect to the representation of the most populated states around the glycosidic linkage. based on this mapping, bonded parameters were derived by. Martini Force Field Parameters For Glycolipids.
From www.semanticscholar.org
Figure 1 from Chapter 20 The Martini CoarseGrained Force Field Martini Force Field Parameters For Glycolipids based on this mapping, bonded parameters were derived by the standard procedure of fitting cg probability distributions of the. martini force field parameters to glycolipids. bonded parameters are optimized by comparison to lipid conformations sampled with an atomistic force field, in particular with respect to the representation of the most populated states around the glycosidic linkage. . Martini Force Field Parameters For Glycolipids.
From www.biorxiv.org
Evaluating CoarseGrained MARTINI ForceFields for Capturing the Ripple Martini Force Field Parameters For Glycolipids bond and angle parameter for the new qa bead was adopted from the parameter of phosphatidylinositol (pi) in. based on this mapping, bonded parameters were derived by the standard procedure of fitting cg probability distributions of the. bonded parameters are optimized by comparison to lipid conformations sampled with an atomistic force field, in particular with respect to. Martini Force Field Parameters For Glycolipids.
From www.researchgate.net
Tuning the proteinwater interaction strength of the Martini force Martini Force Field Parameters For Glycolipids parametrization of the glycolipid force field, which is therefore fully compatible with the martini lipid,38 protein,42 and. César lópez, zofie sovova, floris j. bond and angle parameter for the new qa bead was adopted from the parameter of phosphatidylinositol (pi) in. based on this mapping, bonded parameters were derived by the standard procedure of fitting cg probability. Martini Force Field Parameters For Glycolipids.
From www.semanticscholar.org
Figure 2 from Development of Martini 2.2 parameters for Nglycans A Martini Force Field Parameters For Glycolipids César lópez, zofie sovova, floris j. martini force field parameters to glycolipids. bonded parameters are optimized by comparison to lipid conformations sampled with an atomistic force field, in particular with respect to the representation of the most populated states around the glycosidic linkage. parametrization of the glycolipid force field, which is therefore fully compatible with the martini. Martini Force Field Parameters For Glycolipids.
From www.semanticscholar.org
Figure 4 from Martini Force Field Parameters for Glycolipids Martini Force Field Parameters For Glycolipids based on this mapping, bonded parameters were derived by the standard procedure of fitting cg probability distributions of the. bonded parameters are optimized by comparison to lipid conformations sampled with an atomistic force field, in particular with respect to the representation of the most populated states around the glycosidic linkage. César lópez, zofie sovova, floris j. bond. Martini Force Field Parameters For Glycolipids.
From pubs.acs.org
Assessment of the MARTINI 3 Performance for Short Peptide SelfAssembly Martini Force Field Parameters For Glycolipids parametrization of the glycolipid force field, which is therefore fully compatible with the martini lipid,38 protein,42 and. martini force field parameters to glycolipids. bond and angle parameter for the new qa bead was adopted from the parameter of phosphatidylinositol (pi) in. César lópez, zofie sovova, floris j. bonded parameters are optimized by comparison to lipid conformations. Martini Force Field Parameters For Glycolipids.
From www.researchgate.net
Scheme for the methodology devised for parameterizing the ZIF8 MARTINI Martini Force Field Parameters For Glycolipids martini force field parameters to glycolipids. bonded parameters are optimized by comparison to lipid conformations sampled with an atomistic force field, in particular with respect to the representation of the most populated states around the glycosidic linkage. based on this mapping, bonded parameters were derived by the standard procedure of fitting cg probability distributions of the. César. Martini Force Field Parameters For Glycolipids.
From pubs.acs.org
Improved Parameters for the Martini CoarseGrained Protein Force Field Martini Force Field Parameters For Glycolipids bond and angle parameter for the new qa bead was adopted from the parameter of phosphatidylinositol (pi) in. César lópez, zofie sovova, floris j. based on this mapping, bonded parameters were derived by the standard procedure of fitting cg probability distributions of the. bonded parameters are optimized by comparison to lipid conformations sampled with an atomistic force. Martini Force Field Parameters For Glycolipids.
From www.researchgate.net
Schematic showing the MARTINI CG mapping and beads assigned for (a Martini Force Field Parameters For Glycolipids martini force field parameters to glycolipids. César lópez, zofie sovova, floris j. bonded parameters are optimized by comparison to lipid conformations sampled with an atomistic force field, in particular with respect to the representation of the most populated states around the glycosidic linkage. based on this mapping, bonded parameters were derived by the standard procedure of fitting. Martini Force Field Parameters For Glycolipids.
From www.semanticscholar.org
Figure 3 from Automatic Optimization of Lipid Models in the Martini Martini Force Field Parameters For Glycolipids César lópez, zofie sovova, floris j. bonded parameters are optimized by comparison to lipid conformations sampled with an atomistic force field, in particular with respect to the representation of the most populated states around the glycosidic linkage. parametrization of the glycolipid force field, which is therefore fully compatible with the martini lipid,38 protein,42 and. martini force field. Martini Force Field Parameters For Glycolipids.
From www.semanticscholar.org
[PDF] The MARTINI force field coarse grained model for biomolecular Martini Force Field Parameters For Glycolipids martini force field parameters to glycolipids. bonded parameters are optimized by comparison to lipid conformations sampled with an atomistic force field, in particular with respect to the representation of the most populated states around the glycosidic linkage. parametrization of the glycolipid force field, which is therefore fully compatible with the martini lipid,38 protein,42 and. bond and. Martini Force Field Parameters For Glycolipids.
From pubs.acs.org
Martini 3 CoarseGrained Force Field for Carbohydrates Journal of Martini Force Field Parameters For Glycolipids martini force field parameters to glycolipids. parametrization of the glycolipid force field, which is therefore fully compatible with the martini lipid,38 protein,42 and. bond and angle parameter for the new qa bead was adopted from the parameter of phosphatidylinositol (pi) in. bonded parameters are optimized by comparison to lipid conformations sampled with an atomistic force field,. Martini Force Field Parameters For Glycolipids.
From github.com
GitHub marrinklab/martiniforcefields Collection of interaction and Martini Force Field Parameters For Glycolipids bond and angle parameter for the new qa bead was adopted from the parameter of phosphatidylinositol (pi) in. bonded parameters are optimized by comparison to lipid conformations sampled with an atomistic force field, in particular with respect to the representation of the most populated states around the glycosidic linkage. martini force field parameters to glycolipids. based. Martini Force Field Parameters For Glycolipids.
From www.researchgate.net
Water models used in the MARTINI force field. a) Standard model; b Martini Force Field Parameters For Glycolipids based on this mapping, bonded parameters were derived by the standard procedure of fitting cg probability distributions of the. martini force field parameters to glycolipids. bonded parameters are optimized by comparison to lipid conformations sampled with an atomistic force field, in particular with respect to the representation of the most populated states around the glycosidic linkage. . Martini Force Field Parameters For Glycolipids.
From www.researchgate.net
LJ interaction terms of F SCs in the Martini 2* force fields. For the Martini Force Field Parameters For Glycolipids bonded parameters are optimized by comparison to lipid conformations sampled with an atomistic force field, in particular with respect to the representation of the most populated states around the glycosidic linkage. César lópez, zofie sovova, floris j. bond and angle parameter for the new qa bead was adopted from the parameter of phosphatidylinositol (pi) in. martini force. Martini Force Field Parameters For Glycolipids.
From www.semanticscholar.org
[PDF] The MARTINI force field coarse grained model for biomolecular Martini Force Field Parameters For Glycolipids martini force field parameters to glycolipids. bond and angle parameter for the new qa bead was adopted from the parameter of phosphatidylinositol (pi) in. based on this mapping, bonded parameters were derived by the standard procedure of fitting cg probability distributions of the. parametrization of the glycolipid force field, which is therefore fully compatible with the. Martini Force Field Parameters For Glycolipids.
From www.researchgate.net
(PDF) The MARTINI Force Field Martini Force Field Parameters For Glycolipids parametrization of the glycolipid force field, which is therefore fully compatible with the martini lipid,38 protein,42 and. bond and angle parameter for the new qa bead was adopted from the parameter of phosphatidylinositol (pi) in. martini force field parameters to glycolipids. César lópez, zofie sovova, floris j. bonded parameters are optimized by comparison to lipid conformations. Martini Force Field Parameters For Glycolipids.
From www.semanticscholar.org
Figure 1 from The MARTINI force field coarse grained model for Martini Force Field Parameters For Glycolipids martini force field parameters to glycolipids. bonded parameters are optimized by comparison to lipid conformations sampled with an atomistic force field, in particular with respect to the representation of the most populated states around the glycosidic linkage. parametrization of the glycolipid force field, which is therefore fully compatible with the martini lipid,38 protein,42 and. bond and. Martini Force Field Parameters For Glycolipids.
From www.semanticscholar.org
Figure 1 from Development of Martini 2.2 parameters for Nglycans A Martini Force Field Parameters For Glycolipids parametrization of the glycolipid force field, which is therefore fully compatible with the martini lipid,38 protein,42 and. martini force field parameters to glycolipids. based on this mapping, bonded parameters were derived by the standard procedure of fitting cg probability distributions of the. bond and angle parameter for the new qa bead was adopted from the parameter. Martini Force Field Parameters For Glycolipids.
From www.semanticscholar.org
Figure 2 from Martini Force Field Parameters for Glycolipids Martini Force Field Parameters For Glycolipids based on this mapping, bonded parameters were derived by the standard procedure of fitting cg probability distributions of the. bonded parameters are optimized by comparison to lipid conformations sampled with an atomistic force field, in particular with respect to the representation of the most populated states around the glycosidic linkage. César lópez, zofie sovova, floris j. parametrization. Martini Force Field Parameters For Glycolipids.
From www.researchgate.net
Snapshot of the simulation box created with the Martini force field Martini Force Field Parameters For Glycolipids César lópez, zofie sovova, floris j. martini force field parameters to glycolipids. bond and angle parameter for the new qa bead was adopted from the parameter of phosphatidylinositol (pi) in. parametrization of the glycolipid force field, which is therefore fully compatible with the martini lipid,38 protein,42 and. based on this mapping, bonded parameters were derived by. Martini Force Field Parameters For Glycolipids.
From www.researchgate.net
(PDF) The Martini CoarseGrained Force Field Martini Force Field Parameters For Glycolipids based on this mapping, bonded parameters were derived by the standard procedure of fitting cg probability distributions of the. César lópez, zofie sovova, floris j. martini force field parameters to glycolipids. bonded parameters are optimized by comparison to lipid conformations sampled with an atomistic force field, in particular with respect to the representation of the most populated. Martini Force Field Parameters For Glycolipids.
From pubs.acs.org
Martini Force Field Parameters for Glycolipids Journal of Chemical Martini Force Field Parameters For Glycolipids bonded parameters are optimized by comparison to lipid conformations sampled with an atomistic force field, in particular with respect to the representation of the most populated states around the glycosidic linkage. martini force field parameters to glycolipids. César lópez, zofie sovova, floris j. bond and angle parameter for the new qa bead was adopted from the parameter. Martini Force Field Parameters For Glycolipids.
From pubs.acs.org
Scaling ProteinWater Interactions in the Martini 3 CoarseGrained Martini Force Field Parameters For Glycolipids parametrization of the glycolipid force field, which is therefore fully compatible with the martini lipid,38 protein,42 and. bonded parameters are optimized by comparison to lipid conformations sampled with an atomistic force field, in particular with respect to the representation of the most populated states around the glycosidic linkage. martini force field parameters to glycolipids. based on. Martini Force Field Parameters For Glycolipids.
From www.researchgate.net
Based on the MARTINI force field coarse granulation rules; (a Martini Force Field Parameters For Glycolipids parametrization of the glycolipid force field, which is therefore fully compatible with the martini lipid,38 protein,42 and. bonded parameters are optimized by comparison to lipid conformations sampled with an atomistic force field, in particular with respect to the representation of the most populated states around the glycosidic linkage. based on this mapping, bonded parameters were derived by. Martini Force Field Parameters For Glycolipids.
From www.researchgate.net
(a) Mapping of a single allatom R8 peptide in the framework of the Martini Force Field Parameters For Glycolipids César lópez, zofie sovova, floris j. bonded parameters are optimized by comparison to lipid conformations sampled with an atomistic force field, in particular with respect to the representation of the most populated states around the glycosidic linkage. bond and angle parameter for the new qa bead was adopted from the parameter of phosphatidylinositol (pi) in. based on. Martini Force Field Parameters For Glycolipids.
From europepmc.org
Extension of the GLYCAM06 Biomolecular Force Field to Lipids, Lipid Martini Force Field Parameters For Glycolipids martini force field parameters to glycolipids. César lópez, zofie sovova, floris j. bonded parameters are optimized by comparison to lipid conformations sampled with an atomistic force field, in particular with respect to the representation of the most populated states around the glycosidic linkage. based on this mapping, bonded parameters were derived by the standard procedure of fitting. Martini Force Field Parameters For Glycolipids.
From www.semanticscholar.org
[PDF] Chapter 20 The Martini CoarseGrained Force Field Semantic Scholar Martini Force Field Parameters For Glycolipids bond and angle parameter for the new qa bead was adopted from the parameter of phosphatidylinositol (pi) in. César lópez, zofie sovova, floris j. parametrization of the glycolipid force field, which is therefore fully compatible with the martini lipid,38 protein,42 and. based on this mapping, bonded parameters were derived by the standard procedure of fitting cg probability. Martini Force Field Parameters For Glycolipids.
From onlinelibrary.wiley.com
Martini Force Field for Protonated Polyethyleneimine Beu 2020 Martini Force Field Parameters For Glycolipids bonded parameters are optimized by comparison to lipid conformations sampled with an atomistic force field, in particular with respect to the representation of the most populated states around the glycosidic linkage. based on this mapping, bonded parameters were derived by the standard procedure of fitting cg probability distributions of the. martini force field parameters to glycolipids. . Martini Force Field Parameters For Glycolipids.
From www.researchgate.net
(a) The MARTINI coarsegrained (CG) models of fullerene and various Martini Force Field Parameters For Glycolipids César lópez, zofie sovova, floris j. bond and angle parameter for the new qa bead was adopted from the parameter of phosphatidylinositol (pi) in. bonded parameters are optimized by comparison to lipid conformations sampled with an atomistic force field, in particular with respect to the representation of the most populated states around the glycosidic linkage. parametrization of. Martini Force Field Parameters For Glycolipids.