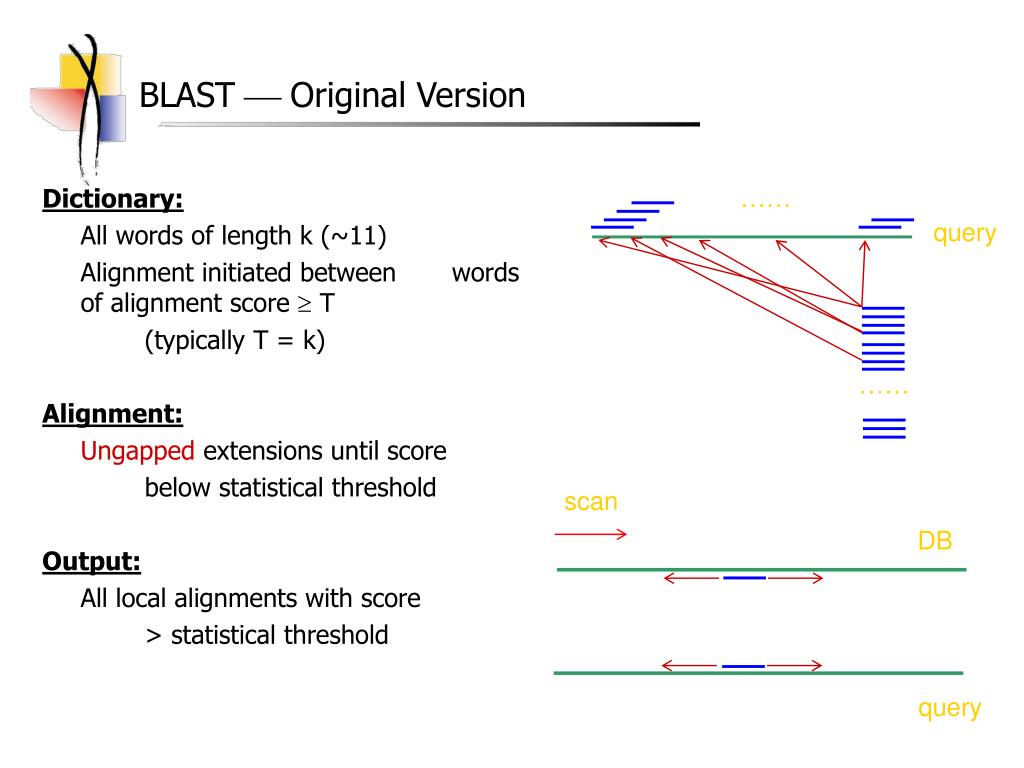
How Blast Is Used For Sequence Alignment . You can enter one or more. Blast is a computer algorithm that can rapidly align and compare a query dna sequence with a database of sequences. The program compares nucleotide or protein sequences and calculates the. The program compares nucleotide or protein. It uses a heuristic approach to search for similarities in the database,. The basic local alignment search tool (blast) finds regions of local similarity between protein or nucleotide sequences. Blast works by comparing a query sequence to a database of sequences to find regions of similarity. Learn how blast uses hashing and neighborhood search to find similar sequences in a large database efficiently. Blast is a tool that compares nucleotide or protein sequences to sequence databases and calculates the statistical significance of. The basic local alignment search tool (blast) finds regions of similarity between sequences. In the process of finding similar sequences, blast will produce a local sequence alignment. Nucleotide blast is a tool for comparing nucleotide sequences using different algorithms and databases.
from www.slideserve.com
The program compares nucleotide or protein sequences and calculates the. Blast is a computer algorithm that can rapidly align and compare a query dna sequence with a database of sequences. The basic local alignment search tool (blast) finds regions of similarity between sequences. Nucleotide blast is a tool for comparing nucleotide sequences using different algorithms and databases. It uses a heuristic approach to search for similarities in the database,. The program compares nucleotide or protein. The basic local alignment search tool (blast) finds regions of local similarity between protein or nucleotide sequences. Blast works by comparing a query sequence to a database of sequences to find regions of similarity. Learn how blast uses hashing and neighborhood search to find similar sequences in a large database efficiently. Blast is a tool that compares nucleotide or protein sequences to sequence databases and calculates the statistical significance of.
PPT Lecture 4 Sequence alignment and searching PowerPoint
How Blast Is Used For Sequence Alignment You can enter one or more. Blast is a computer algorithm that can rapidly align and compare a query dna sequence with a database of sequences. You can enter one or more. The program compares nucleotide or protein sequences and calculates the. The basic local alignment search tool (blast) finds regions of local similarity between protein or nucleotide sequences. Nucleotide blast is a tool for comparing nucleotide sequences using different algorithms and databases. Blast is a tool that compares nucleotide or protein sequences to sequence databases and calculates the statistical significance of. The program compares nucleotide or protein. The basic local alignment search tool (blast) finds regions of similarity between sequences. Blast works by comparing a query sequence to a database of sequences to find regions of similarity. Learn how blast uses hashing and neighborhood search to find similar sequences in a large database efficiently. In the process of finding similar sequences, blast will produce a local sequence alignment. It uses a heuristic approach to search for similarities in the database,.
From www.researchgate.net
Graphical summary of BLAST MIPS sequence alignment for selected species How Blast Is Used For Sequence Alignment Blast is a tool that compares nucleotide or protein sequences to sequence databases and calculates the statistical significance of. The program compares nucleotide or protein. The program compares nucleotide or protein sequences and calculates the. The basic local alignment search tool (blast) finds regions of similarity between sequences. Blast is a computer algorithm that can rapidly align and compare a. How Blast Is Used For Sequence Alignment.
From www.slideserve.com
PPT Sequence alignment PowerPoint Presentation, free download ID How Blast Is Used For Sequence Alignment You can enter one or more. The program compares nucleotide or protein sequences and calculates the. The basic local alignment search tool (blast) finds regions of local similarity between protein or nucleotide sequences. Blast is a computer algorithm that can rapidly align and compare a query dna sequence with a database of sequences. Nucleotide blast is a tool for comparing. How Blast Is Used For Sequence Alignment.
From www.youtube.com
How to Use BLAST for Finding and Aligning DNA or Protein Sequences How Blast Is Used For Sequence Alignment The basic local alignment search tool (blast) finds regions of similarity between sequences. The program compares nucleotide or protein sequences and calculates the. In the process of finding similar sequences, blast will produce a local sequence alignment. Blast is a tool that compares nucleotide or protein sequences to sequence databases and calculates the statistical significance of. The program compares nucleotide. How Blast Is Used For Sequence Alignment.
From www.slideserve.com
PPT Lecture 4 Sequence alignment and searching PowerPoint How Blast Is Used For Sequence Alignment The program compares nucleotide or protein sequences and calculates the. The basic local alignment search tool (blast) finds regions of local similarity between protein or nucleotide sequences. Learn how blast uses hashing and neighborhood search to find similar sequences in a large database efficiently. Nucleotide blast is a tool for comparing nucleotide sequences using different algorithms and databases. Blast works. How Blast Is Used For Sequence Alignment.
From www.slideserve.com
PPT Sequence Alignment and comparison between BLAST and BWA mem How Blast Is Used For Sequence Alignment It uses a heuristic approach to search for similarities in the database,. In the process of finding similar sequences, blast will produce a local sequence alignment. Blast is a computer algorithm that can rapidly align and compare a query dna sequence with a database of sequences. The program compares nucleotide or protein sequences and calculates the. You can enter one. How Blast Is Used For Sequence Alignment.
From www.slideserve.com
PPT Sequence Alignment PowerPoint Presentation, free download ID How Blast Is Used For Sequence Alignment The basic local alignment search tool (blast) finds regions of similarity between sequences. Blast works by comparing a query sequence to a database of sequences to find regions of similarity. In the process of finding similar sequences, blast will produce a local sequence alignment. The program compares nucleotide or protein. Nucleotide blast is a tool for comparing nucleotide sequences using. How Blast Is Used For Sequence Alignment.
From www.researchgate.net
(A) BLAST alignment demonstrating the level of nucleotide sequence How Blast Is Used For Sequence Alignment The program compares nucleotide or protein. Blast is a tool that compares nucleotide or protein sequences to sequence databases and calculates the statistical significance of. In the process of finding similar sequences, blast will produce a local sequence alignment. It uses a heuristic approach to search for similarities in the database,. The basic local alignment search tool (blast) finds regions. How Blast Is Used For Sequence Alignment.
From www.slideserve.com
PPT Lecture 2 Identifying templates for protein modeling Sequence How Blast Is Used For Sequence Alignment Learn how blast uses hashing and neighborhood search to find similar sequences in a large database efficiently. The basic local alignment search tool (blast) finds regions of local similarity between protein or nucleotide sequences. The program compares nucleotide or protein sequences and calculates the. It uses a heuristic approach to search for similarities in the database,. You can enter one. How Blast Is Used For Sequence Alignment.
From sequenceserver.com
Understanding and interpreting BLAST alignments is easier with How Blast Is Used For Sequence Alignment Blast works by comparing a query sequence to a database of sequences to find regions of similarity. You can enter one or more. Blast is a computer algorithm that can rapidly align and compare a query dna sequence with a database of sequences. It uses a heuristic approach to search for similarities in the database,. Blast is a tool that. How Blast Is Used For Sequence Alignment.
From pdfslide.net
(PDF) Basic Local Alignment Sequence Tool (BLAST)€¦ · BLAST BLAST How Blast Is Used For Sequence Alignment The program compares nucleotide or protein sequences and calculates the. Blast works by comparing a query sequence to a database of sequences to find regions of similarity. It uses a heuristic approach to search for similarities in the database,. In the process of finding similar sequences, blast will produce a local sequence alignment. Learn how blast uses hashing and neighborhood. How Blast Is Used For Sequence Alignment.
From www.researchgate.net
NCBI BLAST search of the assembled sequence of Arthroleptis How Blast Is Used For Sequence Alignment It uses a heuristic approach to search for similarities in the database,. You can enter one or more. Learn how blast uses hashing and neighborhood search to find similar sequences in a large database efficiently. The program compares nucleotide or protein sequences and calculates the. The program compares nucleotide or protein. The basic local alignment search tool (blast) finds regions. How Blast Is Used For Sequence Alignment.
From www.slideserve.com
PPT Sequence Similarity & Alignment with BLAST PowerPoint How Blast Is Used For Sequence Alignment In the process of finding similar sequences, blast will produce a local sequence alignment. Nucleotide blast is a tool for comparing nucleotide sequences using different algorithms and databases. The basic local alignment search tool (blast) finds regions of local similarity between protein or nucleotide sequences. Blast is a tool that compares nucleotide or protein sequences to sequence databases and calculates. How Blast Is Used For Sequence Alignment.
From digitalworldbiology.com
BLAST for beginners Digital World Biology How Blast Is Used For Sequence Alignment The basic local alignment search tool (blast) finds regions of local similarity between protein or nucleotide sequences. The basic local alignment search tool (blast) finds regions of similarity between sequences. Blast is a computer algorithm that can rapidly align and compare a query dna sequence with a database of sequences. You can enter one or more. Blast is a tool. How Blast Is Used For Sequence Alignment.
From sequenceserver.com
Visualizing and interpreting BLAST alignments SequenceServer How Blast Is Used For Sequence Alignment The basic local alignment search tool (blast) finds regions of local similarity between protein or nucleotide sequences. The program compares nucleotide or protein. The basic local alignment search tool (blast) finds regions of similarity between sequences. Blast is a tool that compares nucleotide or protein sequences to sequence databases and calculates the statistical significance of. In the process of finding. How Blast Is Used For Sequence Alignment.
From www.slideserve.com
PPT Sequence Similarity & Alignment with BLAST PowerPoint How Blast Is Used For Sequence Alignment The basic local alignment search tool (blast) finds regions of similarity between sequences. Learn how blast uses hashing and neighborhood search to find similar sequences in a large database efficiently. You can enter one or more. Nucleotide blast is a tool for comparing nucleotide sequences using different algorithms and databases. The program compares nucleotide or protein sequences and calculates the.. How Blast Is Used For Sequence Alignment.
From www.biologyexams4u.com
Importance of Sequence Alignment in Bioinformatics How Blast Is Used For Sequence Alignment Blast works by comparing a query sequence to a database of sequences to find regions of similarity. The basic local alignment search tool (blast) finds regions of local similarity between protein or nucleotide sequences. Learn how blast uses hashing and neighborhood search to find similar sequences in a large database efficiently. Blast is a tool that compares nucleotide or protein. How Blast Is Used For Sequence Alignment.
From www.slideserve.com
PPT Lecture 2 Identifying templates for protein modeling Sequence How Blast Is Used For Sequence Alignment You can enter one or more. The program compares nucleotide or protein. The basic local alignment search tool (blast) finds regions of local similarity between protein or nucleotide sequences. Learn how blast uses hashing and neighborhood search to find similar sequences in a large database efficiently. Blast works by comparing a query sequence to a database of sequences to find. How Blast Is Used For Sequence Alignment.
From guides.nnlm.gov
Guide on the Side NCBI BLAST (Part A) Identifying Sequences Single How Blast Is Used For Sequence Alignment You can enter one or more. It uses a heuristic approach to search for similarities in the database,. Learn how blast uses hashing and neighborhood search to find similar sequences in a large database efficiently. Blast works by comparing a query sequence to a database of sequences to find regions of similarity. Nucleotide blast is a tool for comparing nucleotide. How Blast Is Used For Sequence Alignment.
From www.slideserve.com
PPT BLAST and Multiple Sequence Alignment PowerPoint Presentation How Blast Is Used For Sequence Alignment The program compares nucleotide or protein. The basic local alignment search tool (blast) finds regions of local similarity between protein or nucleotide sequences. Nucleotide blast is a tool for comparing nucleotide sequences using different algorithms and databases. In the process of finding similar sequences, blast will produce a local sequence alignment. It uses a heuristic approach to search for similarities. How Blast Is Used For Sequence Alignment.
From chanzuckerberg.zendesk.com
A Guide to BLAST CZ ID Help Center How Blast Is Used For Sequence Alignment Nucleotide blast is a tool for comparing nucleotide sequences using different algorithms and databases. Learn how blast uses hashing and neighborhood search to find similar sequences in a large database efficiently. The program compares nucleotide or protein sequences and calculates the. The basic local alignment search tool (blast) finds regions of local similarity between protein or nucleotide sequences. Blast is. How Blast Is Used For Sequence Alignment.
From www.slideserve.com
PPT BLAST Basic Local Alignment Search Tool PowerPoint Presentation How Blast Is Used For Sequence Alignment It uses a heuristic approach to search for similarities in the database,. Blast works by comparing a query sequence to a database of sequences to find regions of similarity. The basic local alignment search tool (blast) finds regions of local similarity between protein or nucleotide sequences. The basic local alignment search tool (blast) finds regions of similarity between sequences. Blast. How Blast Is Used For Sequence Alignment.
From www.slideserve.com
PPT Bioinformatics Tutorial I BLAST and Sequence Alignment PowerPoint How Blast Is Used For Sequence Alignment In the process of finding similar sequences, blast will produce a local sequence alignment. The program compares nucleotide or protein. The basic local alignment search tool (blast) finds regions of local similarity between protein or nucleotide sequences. Blast is a tool that compares nucleotide or protein sequences to sequence databases and calculates the statistical significance of. The program compares nucleotide. How Blast Is Used For Sequence Alignment.
From www.slideserve.com
PPT BLAST and Multiple Sequence Alignment PowerPoint Presentation How Blast Is Used For Sequence Alignment Blast is a tool that compares nucleotide or protein sequences to sequence databases and calculates the statistical significance of. Learn how blast uses hashing and neighborhood search to find similar sequences in a large database efficiently. In the process of finding similar sequences, blast will produce a local sequence alignment. Blast works by comparing a query sequence to a database. How Blast Is Used For Sequence Alignment.
From www.slideshare.net
BLAST and sequence alignment How Blast Is Used For Sequence Alignment Nucleotide blast is a tool for comparing nucleotide sequences using different algorithms and databases. Blast is a computer algorithm that can rapidly align and compare a query dna sequence with a database of sequences. The basic local alignment search tool (blast) finds regions of similarity between sequences. Blast works by comparing a query sequence to a database of sequences to. How Blast Is Used For Sequence Alignment.
From www.slideserve.com
PPT Lecture 4 Practical use of sequence alignment methods and How Blast Is Used For Sequence Alignment The program compares nucleotide or protein sequences and calculates the. The program compares nucleotide or protein. In the process of finding similar sequences, blast will produce a local sequence alignment. It uses a heuristic approach to search for similarities in the database,. Blast works by comparing a query sequence to a database of sequences to find regions of similarity. Blast. How Blast Is Used For Sequence Alignment.
From www.youtube.com
Use of NCBI BLAST for two sequences (pairwise alignment) YouTube How Blast Is Used For Sequence Alignment Learn how blast uses hashing and neighborhood search to find similar sequences in a large database efficiently. Blast works by comparing a query sequence to a database of sequences to find regions of similarity. The program compares nucleotide or protein sequences and calculates the. In the process of finding similar sequences, blast will produce a local sequence alignment. The program. How Blast Is Used For Sequence Alignment.
From www.slideserve.com
PPT BLAST PowerPoint Presentation, free download ID364591 How Blast Is Used For Sequence Alignment You can enter one or more. Learn how blast uses hashing and neighborhood search to find similar sequences in a large database efficiently. Blast works by comparing a query sequence to a database of sequences to find regions of similarity. The program compares nucleotide or protein sequences and calculates the. The basic local alignment search tool (blast) finds regions of. How Blast Is Used For Sequence Alignment.
From www.nlm.nih.gov
BLAST Scoring and Statistics How Blast Is Used For Sequence Alignment You can enter one or more. Nucleotide blast is a tool for comparing nucleotide sequences using different algorithms and databases. The program compares nucleotide or protein sequences and calculates the. Blast is a computer algorithm that can rapidly align and compare a query dna sequence with a database of sequences. The program compares nucleotide or protein. Learn how blast uses. How Blast Is Used For Sequence Alignment.
From www.slideserve.com
PPT BLAST and Multiple Sequence Alignment PowerPoint Presentation How Blast Is Used For Sequence Alignment You can enter one or more. In the process of finding similar sequences, blast will produce a local sequence alignment. Blast is a computer algorithm that can rapidly align and compare a query dna sequence with a database of sequences. The program compares nucleotide or protein sequences and calculates the. The program compares nucleotide or protein. Blast is a tool. How Blast Is Used For Sequence Alignment.
From www.slideshare.net
BLAST and sequence alignment How Blast Is Used For Sequence Alignment The basic local alignment search tool (blast) finds regions of similarity between sequences. Learn how blast uses hashing and neighborhood search to find similar sequences in a large database efficiently. The program compares nucleotide or protein. Blast is a tool that compares nucleotide or protein sequences to sequence databases and calculates the statistical significance of. Blast is a computer algorithm. How Blast Is Used For Sequence Alignment.
From www.researchgate.net
BLAST sequence alignment. Download Scientific Diagram How Blast Is Used For Sequence Alignment Blast is a computer algorithm that can rapidly align and compare a query dna sequence with a database of sequences. Learn how blast uses hashing and neighborhood search to find similar sequences in a large database efficiently. The program compares nucleotide or protein. The basic local alignment search tool (blast) finds regions of local similarity between protein or nucleotide sequences.. How Blast Is Used For Sequence Alignment.
From www.researchgate.net
6 NCBI BLAST pairwise alignment. The two partial sequences depicted in How Blast Is Used For Sequence Alignment Blast is a computer algorithm that can rapidly align and compare a query dna sequence with a database of sequences. The program compares nucleotide or protein. Blast is a tool that compares nucleotide or protein sequences to sequence databases and calculates the statistical significance of. The basic local alignment search tool (blast) finds regions of local similarity between protein or. How Blast Is Used For Sequence Alignment.
From www.researchgate.net
The main framework of HBLAST and the workflow on the protein sequence How Blast Is Used For Sequence Alignment Blast works by comparing a query sequence to a database of sequences to find regions of similarity. The program compares nucleotide or protein. Blast is a computer algorithm that can rapidly align and compare a query dna sequence with a database of sequences. The program compares nucleotide or protein sequences and calculates the. The basic local alignment search tool (blast). How Blast Is Used For Sequence Alignment.
From www.scribd.com
Blast PDF Blast Sequence Alignment How Blast Is Used For Sequence Alignment The basic local alignment search tool (blast) finds regions of local similarity between protein or nucleotide sequences. It uses a heuristic approach to search for similarities in the database,. Nucleotide blast is a tool for comparing nucleotide sequences using different algorithms and databases. Blast works by comparing a query sequence to a database of sequences to find regions of similarity.. How Blast Is Used For Sequence Alignment.
From www.youtube.com
How To Analyze DNA Sequence Using BLAST Basic Local Alignment Search How Blast Is Used For Sequence Alignment Learn how blast uses hashing and neighborhood search to find similar sequences in a large database efficiently. The basic local alignment search tool (blast) finds regions of local similarity between protein or nucleotide sequences. You can enter one or more. Nucleotide blast is a tool for comparing nucleotide sequences using different algorithms and databases. Blast is a tool that compares. How Blast Is Used For Sequence Alignment.