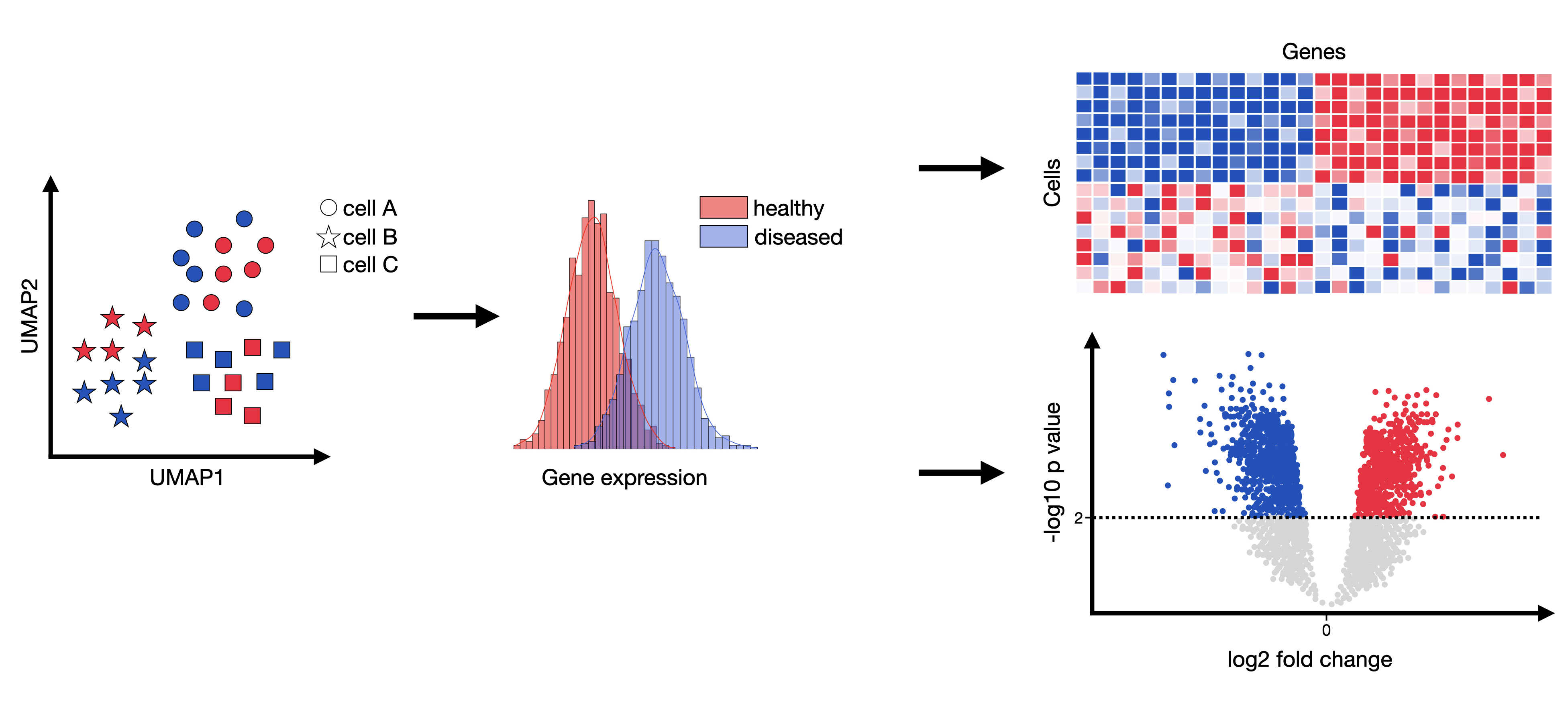
Log Fold Differential Gene Expression . Limma is an r package that was originally developed for differential expression (de) analysis of gene expression microarray data. In r, compute the \(log2\) fold change (“treated. For a given comparison, a positive fold change value indicates an increase of expression, while a negative fold change indicates a decrease in. Specify which groups to compare:. What is the biological meaning of a \(log2\) fold change equal to 1 for gene x? The package deseq2 provides methods to test for differential expression by use of negative binomial generalized linear models;. A log2 (fold change) threshold of 0.5 or 1 is often used to capture relatively small but meaningful.
from www.sc-best-practices.org
For a given comparison, a positive fold change value indicates an increase of expression, while a negative fold change indicates a decrease in. Specify which groups to compare:. What is the biological meaning of a \(log2\) fold change equal to 1 for gene x? In r, compute the \(log2\) fold change (“treated. A log2 (fold change) threshold of 0.5 or 1 is often used to capture relatively small but meaningful. The package deseq2 provides methods to test for differential expression by use of negative binomial generalized linear models;. Limma is an r package that was originally developed for differential expression (de) analysis of gene expression microarray data.
16. Differential gene expression analysis — Singlecell best practices
Log Fold Differential Gene Expression What is the biological meaning of a \(log2\) fold change equal to 1 for gene x? Limma is an r package that was originally developed for differential expression (de) analysis of gene expression microarray data. What is the biological meaning of a \(log2\) fold change equal to 1 for gene x? In r, compute the \(log2\) fold change (“treated. Specify which groups to compare:. The package deseq2 provides methods to test for differential expression by use of negative binomial generalized linear models;. A log2 (fold change) threshold of 0.5 or 1 is often used to capture relatively small but meaningful. For a given comparison, a positive fold change value indicates an increase of expression, while a negative fold change indicates a decrease in.
From www.researchgate.net
Differential expression (Log 2 fold change, LFC) of genes involved in Log Fold Differential Gene Expression In r, compute the \(log2\) fold change (“treated. For a given comparison, a positive fold change value indicates an increase of expression, while a negative fold change indicates a decrease in. What is the biological meaning of a \(log2\) fold change equal to 1 for gene x? Specify which groups to compare:. Limma is an r package that was originally. Log Fold Differential Gene Expression.
From www.researchgate.net
Differential gene expression in the kidney of the wildtype and ΔMtln1 Log Fold Differential Gene Expression For a given comparison, a positive fold change value indicates an increase of expression, while a negative fold change indicates a decrease in. Specify which groups to compare:. A log2 (fold change) threshold of 0.5 or 1 is often used to capture relatively small but meaningful. Limma is an r package that was originally developed for differential expression (de) analysis. Log Fold Differential Gene Expression.
From www.researchgate.net
Differential gene expression analysis for WTand R495Xexpressing Log Fold Differential Gene Expression For a given comparison, a positive fold change value indicates an increase of expression, while a negative fold change indicates a decrease in. Limma is an r package that was originally developed for differential expression (de) analysis of gene expression microarray data. In r, compute the \(log2\) fold change (“treated. What is the biological meaning of a \(log2\) fold change. Log Fold Differential Gene Expression.
From www.vrogue.co
Ma Plot Expression Level Vs Log Fold Change Of Differ vrogue.co Log Fold Differential Gene Expression Limma is an r package that was originally developed for differential expression (de) analysis of gene expression microarray data. A log2 (fold change) threshold of 0.5 or 1 is often used to capture relatively small but meaningful. Specify which groups to compare:. In r, compute the \(log2\) fold change (“treated. For a given comparison, a positive fold change value indicates. Log Fold Differential Gene Expression.
From biocorecrg.github.io
Differential gene expression Log Fold Differential Gene Expression What is the biological meaning of a \(log2\) fold change equal to 1 for gene x? In r, compute the \(log2\) fold change (“treated. For a given comparison, a positive fold change value indicates an increase of expression, while a negative fold change indicates a decrease in. Specify which groups to compare:. Limma is an r package that was originally. Log Fold Differential Gene Expression.
From www.researchgate.net
ab Volcano plots for differential gene expression (average log fold Log Fold Differential Gene Expression Limma is an r package that was originally developed for differential expression (de) analysis of gene expression microarray data. For a given comparison, a positive fold change value indicates an increase of expression, while a negative fold change indicates a decrease in. What is the biological meaning of a \(log2\) fold change equal to 1 for gene x? A log2. Log Fold Differential Gene Expression.
From www.researchgate.net
Differential gene expression identifies modules that contains Log Fold Differential Gene Expression For a given comparison, a positive fold change value indicates an increase of expression, while a negative fold change indicates a decrease in. Specify which groups to compare:. The package deseq2 provides methods to test for differential expression by use of negative binomial generalized linear models;. What is the biological meaning of a \(log2\) fold change equal to 1 for. Log Fold Differential Gene Expression.
From www.researchgate.net
Pearson correlation of gene expression fold changes and differential Log Fold Differential Gene Expression In r, compute the \(log2\) fold change (“treated. What is the biological meaning of a \(log2\) fold change equal to 1 for gene x? A log2 (fold change) threshold of 0.5 or 1 is often used to capture relatively small but meaningful. The package deseq2 provides methods to test for differential expression by use of negative binomial generalized linear models;.. Log Fold Differential Gene Expression.
From www.vrogue.co
Ma Plot Expression Level Vs Log Fold Change Of Differ vrogue.co Log Fold Differential Gene Expression Limma is an r package that was originally developed for differential expression (de) analysis of gene expression microarray data. What is the biological meaning of a \(log2\) fold change equal to 1 for gene x? Specify which groups to compare:. A log2 (fold change) threshold of 0.5 or 1 is often used to capture relatively small but meaningful. In r,. Log Fold Differential Gene Expression.
From www.researchgate.net
Volcano plots showing differential expression in log 2 fold change on Log Fold Differential Gene Expression Limma is an r package that was originally developed for differential expression (de) analysis of gene expression microarray data. For a given comparison, a positive fold change value indicates an increase of expression, while a negative fold change indicates a decrease in. The package deseq2 provides methods to test for differential expression by use of negative binomial generalized linear models;.. Log Fold Differential Gene Expression.
From www.researchgate.net
Differential gene expression (log 2 foldchange) of virulence genes Log Fold Differential Gene Expression Specify which groups to compare:. The package deseq2 provides methods to test for differential expression by use of negative binomial generalized linear models;. Limma is an r package that was originally developed for differential expression (de) analysis of gene expression microarray data. A log2 (fold change) threshold of 0.5 or 1 is often used to capture relatively small but meaningful.. Log Fold Differential Gene Expression.
From www.researchgate.net
Validation of RNASeq differential gene expression results. Log 2 Log Fold Differential Gene Expression For a given comparison, a positive fold change value indicates an increase of expression, while a negative fold change indicates a decrease in. Limma is an r package that was originally developed for differential expression (de) analysis of gene expression microarray data. What is the biological meaning of a \(log2\) fold change equal to 1 for gene x? A log2. Log Fold Differential Gene Expression.
From www.researchgate.net
Differential expression (i.e., log fold change) by larvae within low pH Log Fold Differential Gene Expression A log2 (fold change) threshold of 0.5 or 1 is often used to capture relatively small but meaningful. For a given comparison, a positive fold change value indicates an increase of expression, while a negative fold change indicates a decrease in. In r, compute the \(log2\) fold change (“treated. What is the biological meaning of a \(log2\) fold change equal. Log Fold Differential Gene Expression.
From www.researchgate.net
Heatmap of logfold change (LFC) comparisons of differential gene Log Fold Differential Gene Expression In r, compute the \(log2\) fold change (“treated. Specify which groups to compare:. Limma is an r package that was originally developed for differential expression (de) analysis of gene expression microarray data. The package deseq2 provides methods to test for differential expression by use of negative binomial generalized linear models;. A log2 (fold change) threshold of 0.5 or 1 is. Log Fold Differential Gene Expression.
From www.researchgate.net
Differential gene expression during development. Volcano plots (log2 Log Fold Differential Gene Expression In r, compute the \(log2\) fold change (“treated. A log2 (fold change) threshold of 0.5 or 1 is often used to capture relatively small but meaningful. Limma is an r package that was originally developed for differential expression (de) analysis of gene expression microarray data. Specify which groups to compare:. What is the biological meaning of a \(log2\) fold change. Log Fold Differential Gene Expression.
From www.researchgate.net
Differential gene expression of memoryrelated genes. Log2 fold changes Log Fold Differential Gene Expression What is the biological meaning of a \(log2\) fold change equal to 1 for gene x? For a given comparison, a positive fold change value indicates an increase of expression, while a negative fold change indicates a decrease in. In r, compute the \(log2\) fold change (“treated. Limma is an r package that was originally developed for differential expression (de). Log Fold Differential Gene Expression.
From www.researchgate.net
Volcano plot showing variation of significance and logfoldchange for Log Fold Differential Gene Expression Specify which groups to compare:. The package deseq2 provides methods to test for differential expression by use of negative binomial generalized linear models;. In r, compute the \(log2\) fold change (“treated. What is the biological meaning of a \(log2\) fold change equal to 1 for gene x? A log2 (fold change) threshold of 0.5 or 1 is often used to. Log Fold Differential Gene Expression.
From www.researchgate.net
MDplot showing the logfold change of differential expressed lncRNAs Log Fold Differential Gene Expression Limma is an r package that was originally developed for differential expression (de) analysis of gene expression microarray data. For a given comparison, a positive fold change value indicates an increase of expression, while a negative fold change indicates a decrease in. The package deseq2 provides methods to test for differential expression by use of negative binomial generalized linear models;.. Log Fold Differential Gene Expression.
From www.researchgate.net
Box plots of log fold changes of normalized gene expression from Log Fold Differential Gene Expression Limma is an r package that was originally developed for differential expression (de) analysis of gene expression microarray data. A log2 (fold change) threshold of 0.5 or 1 is often used to capture relatively small but meaningful. The package deseq2 provides methods to test for differential expression by use of negative binomial generalized linear models;. For a given comparison, a. Log Fold Differential Gene Expression.
From www.researchgate.net
(A) Differential gene MA map. The ordinate represents the log2 fold Log Fold Differential Gene Expression Specify which groups to compare:. What is the biological meaning of a \(log2\) fold change equal to 1 for gene x? In r, compute the \(log2\) fold change (“treated. For a given comparison, a positive fold change value indicates an increase of expression, while a negative fold change indicates a decrease in. A log2 (fold change) threshold of 0.5 or. Log Fold Differential Gene Expression.
From www.researchgate.net
Log foldchange in gene expression relative to iPSCs. (A) Primary Log Fold Differential Gene Expression What is the biological meaning of a \(log2\) fold change equal to 1 for gene x? Specify which groups to compare:. For a given comparison, a positive fold change value indicates an increase of expression, while a negative fold change indicates a decrease in. The package deseq2 provides methods to test for differential expression by use of negative binomial generalized. Log Fold Differential Gene Expression.
From www.researchgate.net
Heatmap showing log2 fold change to visualize gene expression Log Fold Differential Gene Expression The package deseq2 provides methods to test for differential expression by use of negative binomial generalized linear models;. A log2 (fold change) threshold of 0.5 or 1 is often used to capture relatively small but meaningful. Specify which groups to compare:. For a given comparison, a positive fold change value indicates an increase of expression, while a negative fold change. Log Fold Differential Gene Expression.
From www.researchgate.net
Differential gene expression and KEGG analysis of IEC6 cells. a Two Log Fold Differential Gene Expression Limma is an r package that was originally developed for differential expression (de) analysis of gene expression microarray data. For a given comparison, a positive fold change value indicates an increase of expression, while a negative fold change indicates a decrease in. In r, compute the \(log2\) fold change (“treated. What is the biological meaning of a \(log2\) fold change. Log Fold Differential Gene Expression.
From www.researchgate.net
Differential gene expression analyses by edgeR and validation of Log Fold Differential Gene Expression For a given comparison, a positive fold change value indicates an increase of expression, while a negative fold change indicates a decrease in. In r, compute the \(log2\) fold change (“treated. The package deseq2 provides methods to test for differential expression by use of negative binomial generalized linear models;. Specify which groups to compare:. Limma is an r package that. Log Fold Differential Gene Expression.
From www.researchgate.net
Differential gene expression analysis. UpSetR [78] plots highlighting Log Fold Differential Gene Expression Limma is an r package that was originally developed for differential expression (de) analysis of gene expression microarray data. In r, compute the \(log2\) fold change (“treated. For a given comparison, a positive fold change value indicates an increase of expression, while a negative fold change indicates a decrease in. A log2 (fold change) threshold of 0.5 or 1 is. Log Fold Differential Gene Expression.
From www.researchgate.net
Heatmap of log 2 foldchange value of differently expression genes in Log Fold Differential Gene Expression A log2 (fold change) threshold of 0.5 or 1 is often used to capture relatively small but meaningful. Specify which groups to compare:. In r, compute the \(log2\) fold change (“treated. Limma is an r package that was originally developed for differential expression (de) analysis of gene expression microarray data. For a given comparison, a positive fold change value indicates. Log Fold Differential Gene Expression.
From www.vrogue.co
Ma Plot Expression Level Vs Log Fold Change Of Differ vrogue.co Log Fold Differential Gene Expression In r, compute the \(log2\) fold change (“treated. What is the biological meaning of a \(log2\) fold change equal to 1 for gene x? The package deseq2 provides methods to test for differential expression by use of negative binomial generalized linear models;. For a given comparison, a positive fold change value indicates an increase of expression, while a negative fold. Log Fold Differential Gene Expression.
From www.researchgate.net
Differential gene expression plot. The fold change in the expression of Log Fold Differential Gene Expression A log2 (fold change) threshold of 0.5 or 1 is often used to capture relatively small but meaningful. For a given comparison, a positive fold change value indicates an increase of expression, while a negative fold change indicates a decrease in. Limma is an r package that was originally developed for differential expression (de) analysis of gene expression microarray data.. Log Fold Differential Gene Expression.
From www.researchgate.net
Overall interplatform gene expression analysis as fold change for each Log Fold Differential Gene Expression For a given comparison, a positive fold change value indicates an increase of expression, while a negative fold change indicates a decrease in. What is the biological meaning of a \(log2\) fold change equal to 1 for gene x? In r, compute the \(log2\) fold change (“treated. A log2 (fold change) threshold of 0.5 or 1 is often used to. Log Fold Differential Gene Expression.
From www.researchgate.net
Bean plots of differential expression levels (logfold change) of Log Fold Differential Gene Expression The package deseq2 provides methods to test for differential expression by use of negative binomial generalized linear models;. Specify which groups to compare:. For a given comparison, a positive fold change value indicates an increase of expression, while a negative fold change indicates a decrease in. Limma is an r package that was originally developed for differential expression (de) analysis. Log Fold Differential Gene Expression.
From www.researchgate.net
Differential gene expression analysis by subtype. Venn diagram Log Fold Differential Gene Expression A log2 (fold change) threshold of 0.5 or 1 is often used to capture relatively small but meaningful. For a given comparison, a positive fold change value indicates an increase of expression, while a negative fold change indicates a decrease in. Limma is an r package that was originally developed for differential expression (de) analysis of gene expression microarray data.. Log Fold Differential Gene Expression.
From www.researchgate.net
Heatmap of logfold change (LFC) comparisons of differential gene Log Fold Differential Gene Expression Limma is an r package that was originally developed for differential expression (de) analysis of gene expression microarray data. A log2 (fold change) threshold of 0.5 or 1 is often used to capture relatively small but meaningful. For a given comparison, a positive fold change value indicates an increase of expression, while a negative fold change indicates a decrease in.. Log Fold Differential Gene Expression.
From www.researchgate.net
Differential gene expression during early heterokaryon... Download Log Fold Differential Gene Expression A log2 (fold change) threshold of 0.5 or 1 is often used to capture relatively small but meaningful. In r, compute the \(log2\) fold change (“treated. The package deseq2 provides methods to test for differential expression by use of negative binomial generalized linear models;. Limma is an r package that was originally developed for differential expression (de) analysis of gene. Log Fold Differential Gene Expression.
From www.researchgate.net
Differential gene expression at transcriptional and translational Log Fold Differential Gene Expression A log2 (fold change) threshold of 0.5 or 1 is often used to capture relatively small but meaningful. The package deseq2 provides methods to test for differential expression by use of negative binomial generalized linear models;. In r, compute the \(log2\) fold change (“treated. What is the biological meaning of a \(log2\) fold change equal to 1 for gene x?. Log Fold Differential Gene Expression.
From www.sc-best-practices.org
16. Differential gene expression analysis — Singlecell best practices Log Fold Differential Gene Expression Specify which groups to compare:. In r, compute the \(log2\) fold change (“treated. What is the biological meaning of a \(log2\) fold change equal to 1 for gene x? The package deseq2 provides methods to test for differential expression by use of negative binomial generalized linear models;. A log2 (fold change) threshold of 0.5 or 1 is often used to. Log Fold Differential Gene Expression.