Protein Folding Server . (1) using the alphafold2 model with residue index jump and (2) using the. The orion web server accepts a single protein sequence as input and searches homologous protein structures within minutes. Alphamissense leverages alphafold2’s capability to model protein structure, and its capacity to learn evolutionary constraints from related. The phyre automatic fold recognition server for predicting the structure and/or function of your protein sequence. View over 200 million protein structure predictions to support your research. Phyre standard mode for job. Raptorx predicts protein secondary and tertiary structures, contact and distance map, solvent accessibility, disordered regions, functional. Colabfold is a free and accessible platform for protein folding that provides accelerated prediction of protein structures and. We currently have two different ways to predict protein complexes: Alphafold server predict the structure and interactions of all life’s.
from www.linkedin.com
(1) using the alphafold2 model with residue index jump and (2) using the. The phyre automatic fold recognition server for predicting the structure and/or function of your protein sequence. Alphafold server predict the structure and interactions of all life’s. The orion web server accepts a single protein sequence as input and searches homologous protein structures within minutes. Alphamissense leverages alphafold2’s capability to model protein structure, and its capacity to learn evolutionary constraints from related. Colabfold is a free and accessible platform for protein folding that provides accelerated prediction of protein structures and. Raptorx predicts protein secondary and tertiary structures, contact and distance map, solvent accessibility, disordered regions, functional. We currently have two different ways to predict protein complexes: View over 200 million protein structure predictions to support your research. Phyre standard mode for job.
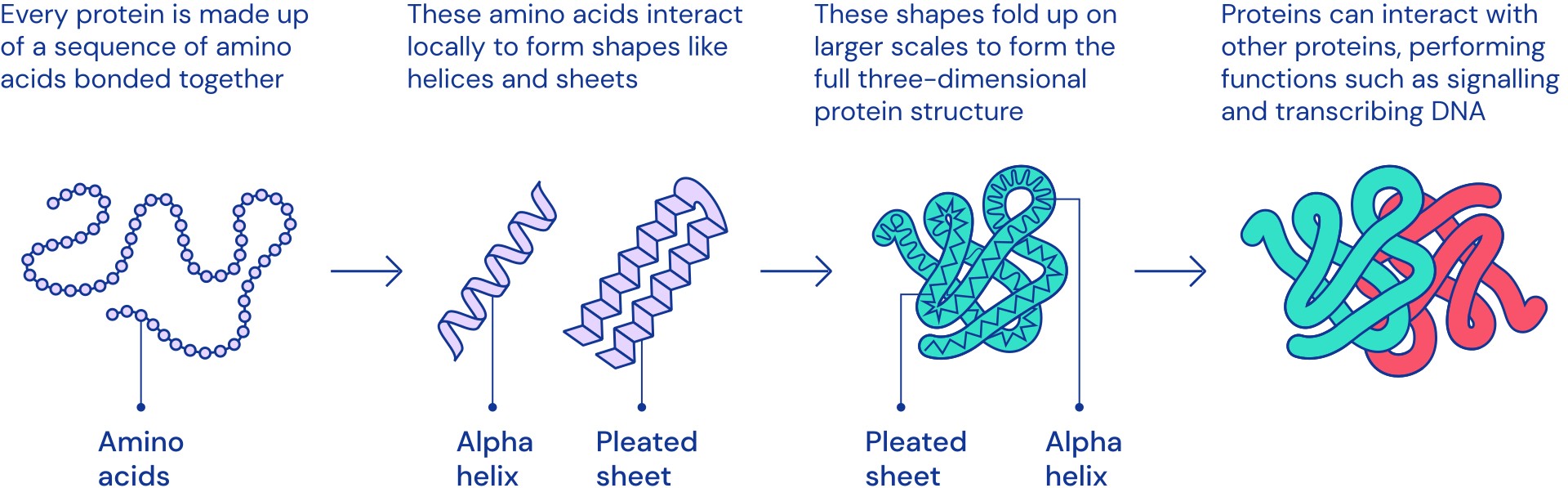
Why is protein folding such a big deal?
Protein Folding Server The phyre automatic fold recognition server for predicting the structure and/or function of your protein sequence. Colabfold is a free and accessible platform for protein folding that provides accelerated prediction of protein structures and. The phyre automatic fold recognition server for predicting the structure and/or function of your protein sequence. Phyre standard mode for job. View over 200 million protein structure predictions to support your research. We currently have two different ways to predict protein complexes: Raptorx predicts protein secondary and tertiary structures, contact and distance map, solvent accessibility, disordered regions, functional. The orion web server accepts a single protein sequence as input and searches homologous protein structures within minutes. Alphafold server predict the structure and interactions of all life’s. (1) using the alphafold2 model with residue index jump and (2) using the. Alphamissense leverages alphafold2’s capability to model protein structure, and its capacity to learn evolutionary constraints from related.
From atlasbars.com
Protein Folding Exploring the Intricate Process of Protein Folding and Protein Folding Server We currently have two different ways to predict protein complexes: (1) using the alphafold2 model with residue index jump and (2) using the. View over 200 million protein structure predictions to support your research. Alphamissense leverages alphafold2’s capability to model protein structure, and its capacity to learn evolutionary constraints from related. The orion web server accepts a single protein sequence. Protein Folding Server.
From www.youtube.com
DeepMind AlphaFold2 Protein Folding tutorial on Google Colab python Protein Folding Server Colabfold is a free and accessible platform for protein folding that provides accelerated prediction of protein structures and. Phyre standard mode for job. The orion web server accepts a single protein sequence as input and searches homologous protein structures within minutes. Alphamissense leverages alphafold2’s capability to model protein structure, and its capacity to learn evolutionary constraints from related. (1) using. Protein Folding Server.
From www.sbg.bio.ic.ac.uk
PHYRE Protein Fold Recognition Server Protein Folding Server (1) using the alphafold2 model with residue index jump and (2) using the. Phyre standard mode for job. View over 200 million protein structure predictions to support your research. Alphamissense leverages alphafold2’s capability to model protein structure, and its capacity to learn evolutionary constraints from related. We currently have two different ways to predict protein complexes: Raptorx predicts protein secondary. Protein Folding Server.
From www.readkong.com
BESTSEL WEBSERVER FOR SECONDARY STRUCTURE AND FOLD PREDICTION FOR Protein Folding Server Colabfold is a free and accessible platform for protein folding that provides accelerated prediction of protein structures and. Raptorx predicts protein secondary and tertiary structures, contact and distance map, solvent accessibility, disordered regions, functional. Phyre standard mode for job. (1) using the alphafold2 model with residue index jump and (2) using the. Alphamissense leverages alphafold2’s capability to model protein structure,. Protein Folding Server.
From www.researchgate.net
(PDF) ORION A web server for protein fold recognition and structure Protein Folding Server Alphamissense leverages alphafold2’s capability to model protein structure, and its capacity to learn evolutionary constraints from related. (1) using the alphafold2 model with residue index jump and (2) using the. The phyre automatic fold recognition server for predicting the structure and/or function of your protein sequence. Colabfold is a free and accessible platform for protein folding that provides accelerated prediction. Protein Folding Server.
From quantumzeitgeist.com
Protein Folding Takes A Step Forward With Quantum Computing Protein Folding Server Raptorx predicts protein secondary and tertiary structures, contact and distance map, solvent accessibility, disordered regions, functional. Alphamissense leverages alphafold2’s capability to model protein structure, and its capacity to learn evolutionary constraints from related. Phyre standard mode for job. The phyre automatic fold recognition server for predicting the structure and/or function of your protein sequence. Colabfold is a free and accessible. Protein Folding Server.
From www.linkedin.com
Why is protein folding such a big deal? Protein Folding Server The orion web server accepts a single protein sequence as input and searches homologous protein structures within minutes. Phyre standard mode for job. View over 200 million protein structure predictions to support your research. Colabfold is a free and accessible platform for protein folding that provides accelerated prediction of protein structures and. Alphamissense leverages alphafold2’s capability to model protein structure,. Protein Folding Server.
From www.slideserve.com
PPT Protein Folding Protein Structure Prediction Protein Design Protein Folding Server View over 200 million protein structure predictions to support your research. We currently have two different ways to predict protein complexes: Raptorx predicts protein secondary and tertiary structures, contact and distance map, solvent accessibility, disordered regions, functional. Alphafold server predict the structure and interactions of all life’s. Phyre standard mode for job. Alphamissense leverages alphafold2’s capability to model protein structure,. Protein Folding Server.
From www.researchgate.net
Hierarchical protein folding. (A) Progression of a protein folding Protein Folding Server Phyre standard mode for job. Raptorx predicts protein secondary and tertiary structures, contact and distance map, solvent accessibility, disordered regions, functional. Alphafold server predict the structure and interactions of all life’s. Colabfold is a free and accessible platform for protein folding that provides accelerated prediction of protein structures and. View over 200 million protein structure predictions to support your research.. Protein Folding Server.
From www.researchgate.net
0.3 Reverse engineering the protein folding process for protein design Protein Folding Server Colabfold is a free and accessible platform for protein folding that provides accelerated prediction of protein structures and. Phyre standard mode for job. The phyre automatic fold recognition server for predicting the structure and/or function of your protein sequence. Raptorx predicts protein secondary and tertiary structures, contact and distance map, solvent accessibility, disordered regions, functional. We currently have two different. Protein Folding Server.
From www.youtube.com
Threading/ Fold recognition through Phyr2 Server and Abinitio Protein Protein Folding Server View over 200 million protein structure predictions to support your research. Raptorx predicts protein secondary and tertiary structures, contact and distance map, solvent accessibility, disordered regions, functional. Phyre standard mode for job. We currently have two different ways to predict protein complexes: The orion web server accepts a single protein sequence as input and searches homologous protein structures within minutes.. Protein Folding Server.
From www.researchgate.net
Modeling of protein folding by the RaptorX web server (Källberg et al Protein Folding Server Raptorx predicts protein secondary and tertiary structures, contact and distance map, solvent accessibility, disordered regions, functional. Alphamissense leverages alphafold2’s capability to model protein structure, and its capacity to learn evolutionary constraints from related. Alphafold server predict the structure and interactions of all life’s. We currently have two different ways to predict protein complexes: (1) using the alphafold2 model with residue. Protein Folding Server.
From www.researchgate.net
(PDF) FoldRate A Server for Predicting Protein Folding Rates from Protein Folding Server Colabfold is a free and accessible platform for protein folding that provides accelerated prediction of protein structures and. Raptorx predicts protein secondary and tertiary structures, contact and distance map, solvent accessibility, disordered regions, functional. Phyre standard mode for job. The phyre automatic fold recognition server for predicting the structure and/or function of your protein sequence. We currently have two different. Protein Folding Server.
From www.researchgate.net
Reconstructed MEP networks for proteinfolding transitions. We compare Protein Folding Server Phyre standard mode for job. View over 200 million protein structure predictions to support your research. Colabfold is a free and accessible platform for protein folding that provides accelerated prediction of protein structures and. The phyre automatic fold recognition server for predicting the structure and/or function of your protein sequence. We currently have two different ways to predict protein complexes:. Protein Folding Server.
From automatski.com
Protein Folding Protein Folding Server Phyre standard mode for job. Alphafold server predict the structure and interactions of all life’s. Alphamissense leverages alphafold2’s capability to model protein structure, and its capacity to learn evolutionary constraints from related. We currently have two different ways to predict protein complexes: The orion web server accepts a single protein sequence as input and searches homologous protein structures within minutes.. Protein Folding Server.
From www.photophysics.com
Protein Folding Protein Folding Server Phyre standard mode for job. View over 200 million protein structure predictions to support your research. Alphamissense leverages alphafold2’s capability to model protein structure, and its capacity to learn evolutionary constraints from related. (1) using the alphafold2 model with residue index jump and (2) using the. The phyre automatic fold recognition server for predicting the structure and/or function of your. Protein Folding Server.
From www.researchgate.net
Proteinprotein web network for 10 fold Upregulated proteins in Protein Folding Server Phyre standard mode for job. Alphamissense leverages alphafold2’s capability to model protein structure, and its capacity to learn evolutionary constraints from related. The orion web server accepts a single protein sequence as input and searches homologous protein structures within minutes. Alphafold server predict the structure and interactions of all life’s. View over 200 million protein structure predictions to support your. Protein Folding Server.
From slidetodoc.com
Protein Folding PROTEIN FOLDING Process in which a Protein Folding Server Raptorx predicts protein secondary and tertiary structures, contact and distance map, solvent accessibility, disordered regions, functional. The orion web server accepts a single protein sequence as input and searches homologous protein structures within minutes. Alphamissense leverages alphafold2’s capability to model protein structure, and its capacity to learn evolutionary constraints from related. Alphafold server predict the structure and interactions of all. Protein Folding Server.
From news.rice.edu
Folding proteins feel the heat, and cold Rice News News and Media Protein Folding Server (1) using the alphafold2 model with residue index jump and (2) using the. Raptorx predicts protein secondary and tertiary structures, contact and distance map, solvent accessibility, disordered regions, functional. The phyre automatic fold recognition server for predicting the structure and/or function of your protein sequence. Colabfold is a free and accessible platform for protein folding that provides accelerated prediction of. Protein Folding Server.
From analyticsindiamag.com
Unfolding Protein Folding With ESM2 Protein Folding Server The orion web server accepts a single protein sequence as input and searches homologous protein structures within minutes. Alphamissense leverages alphafold2’s capability to model protein structure, and its capacity to learn evolutionary constraints from related. View over 200 million protein structure predictions to support your research. (1) using the alphafold2 model with residue index jump and (2) using the. We. Protein Folding Server.
From proteinchoices.blogspot.com
Protein folding Refolding Proteins Protein Choices Protein Folding Server Alphamissense leverages alphafold2’s capability to model protein structure, and its capacity to learn evolutionary constraints from related. The phyre automatic fold recognition server for predicting the structure and/or function of your protein sequence. Raptorx predicts protein secondary and tertiary structures, contact and distance map, solvent accessibility, disordered regions, functional. Alphafold server predict the structure and interactions of all life’s. The. Protein Folding Server.
From www.researchgate.net
(PDF) TMFold A web server for predicting transmembrane protein fold Protein Folding Server (1) using the alphafold2 model with residue index jump and (2) using the. Colabfold is a free and accessible platform for protein folding that provides accelerated prediction of protein structures and. View over 200 million protein structure predictions to support your research. The orion web server accepts a single protein sequence as input and searches homologous protein structures within minutes.. Protein Folding Server.
From aws.amazon.com
Guidance for Protein Folding on AWS Protein Folding Server We currently have two different ways to predict protein complexes: Alphamissense leverages alphafold2’s capability to model protein structure, and its capacity to learn evolutionary constraints from related. Phyre standard mode for job. Alphafold server predict the structure and interactions of all life’s. Colabfold is a free and accessible platform for protein folding that provides accelerated prediction of protein structures and.. Protein Folding Server.
From www.pnas.org
De novo protein fold design through sequenceindependent fragment Protein Folding Server Alphafold server predict the structure and interactions of all life’s. Raptorx predicts protein secondary and tertiary structures, contact and distance map, solvent accessibility, disordered regions, functional. The phyre automatic fold recognition server for predicting the structure and/or function of your protein sequence. We currently have two different ways to predict protein complexes: The orion web server accepts a single protein. Protein Folding Server.
From upmytech.com
AI for protein folding [MIT Tech Review] Up My Tech Protein Folding Server Alphamissense leverages alphafold2’s capability to model protein structure, and its capacity to learn evolutionary constraints from related. Colabfold is a free and accessible platform for protein folding that provides accelerated prediction of protein structures and. We currently have two different ways to predict protein complexes: The phyre automatic fold recognition server for predicting the structure and/or function of your protein. Protein Folding Server.
From www.youtube.com
Protein folding explained YouTube Protein Folding Server Alphamissense leverages alphafold2’s capability to model protein structure, and its capacity to learn evolutionary constraints from related. Phyre standard mode for job. Colabfold is a free and accessible platform for protein folding that provides accelerated prediction of protein structures and. Alphafold server predict the structure and interactions of all life’s. View over 200 million protein structure predictions to support your. Protein Folding Server.
From www.slideserve.com
PPT Protein Folding and Processing PowerPoint Presentation, free Protein Folding Server The phyre automatic fold recognition server for predicting the structure and/or function of your protein sequence. Alphafold server predict the structure and interactions of all life’s. Raptorx predicts protein secondary and tertiary structures, contact and distance map, solvent accessibility, disordered regions, functional. (1) using the alphafold2 model with residue index jump and (2) using the. View over 200 million protein. Protein Folding Server.
From gamma.app
Protein Folding Gamma Protein Folding Server (1) using the alphafold2 model with residue index jump and (2) using the. Alphafold server predict the structure and interactions of all life’s. Colabfold is a free and accessible platform for protein folding that provides accelerated prediction of protein structures and. The orion web server accepts a single protein sequence as input and searches homologous protein structures within minutes. Phyre. Protein Folding Server.
From www.science.org
How FastFolding Proteins Fold Science Protein Folding Server Phyre standard mode for job. (1) using the alphafold2 model with residue index jump and (2) using the. The orion web server accepts a single protein sequence as input and searches homologous protein structures within minutes. Alphafold server predict the structure and interactions of all life’s. Alphamissense leverages alphafold2’s capability to model protein structure, and its capacity to learn evolutionary. Protein Folding Server.
From www.ebi.ac.uk
Fold Biomacromolecular structures Protein Folding Server The phyre automatic fold recognition server for predicting the structure and/or function of your protein sequence. The orion web server accepts a single protein sequence as input and searches homologous protein structures within minutes. We currently have two different ways to predict protein complexes: Alphamissense leverages alphafold2’s capability to model protein structure, and its capacity to learn evolutionary constraints from. Protein Folding Server.
From www.researchgate.net
2 The metaserver approach for protein structure prediction. The Protein Folding Server Colabfold is a free and accessible platform for protein folding that provides accelerated prediction of protein structures and. View over 200 million protein structure predictions to support your research. The orion web server accepts a single protein sequence as input and searches homologous protein structures within minutes. We currently have two different ways to predict protein complexes: Alphamissense leverages alphafold2’s. Protein Folding Server.
From analyticsdrift.com
Top 5 protein folding models Analytics Drift Data Science Protein Folding Server Alphafold server predict the structure and interactions of all life’s. The orion web server accepts a single protein sequence as input and searches homologous protein structures within minutes. We currently have two different ways to predict protein complexes: Raptorx predicts protein secondary and tertiary structures, contact and distance map, solvent accessibility, disordered regions, functional. (1) using the alphafold2 model with. Protein Folding Server.
From phys.org
Folding proteins feel the heat, and cold Protein Folding Server View over 200 million protein structure predictions to support your research. Alphamissense leverages alphafold2’s capability to model protein structure, and its capacity to learn evolutionary constraints from related. Phyre standard mode for job. We currently have two different ways to predict protein complexes: Colabfold is a free and accessible platform for protein folding that provides accelerated prediction of protein structures. Protein Folding Server.
From www.science.org
How FastFolding Proteins Fold Science Protein Folding Server Alphamissense leverages alphafold2’s capability to model protein structure, and its capacity to learn evolutionary constraints from related. Phyre standard mode for job. Raptorx predicts protein secondary and tertiary structures, contact and distance map, solvent accessibility, disordered regions, functional. View over 200 million protein structure predictions to support your research. (1) using the alphafold2 model with residue index jump and (2). Protein Folding Server.
From www.kroll-software.ch
Protein Folding by a Neural Network KROLLSOFTWARE Protein Folding Server Phyre standard mode for job. (1) using the alphafold2 model with residue index jump and (2) using the. Alphamissense leverages alphafold2’s capability to model protein structure, and its capacity to learn evolutionary constraints from related. View over 200 million protein structure predictions to support your research. Raptorx predicts protein secondary and tertiary structures, contact and distance map, solvent accessibility, disordered. Protein Folding Server.