Haddock Antibody . We find that haddock can generate accurate models of antibodyantigen complexes using an ensemble of antibody. We use anarci [dunbar, j. Here we provide the code to run the antibody protocol of haddock by using the residues belonging to the hypervariable (hv) loops. Haddock is a method for modeling biomolecular complexes using ambiguous interaction restraints and other experimental data. To present the web server’s various interfaces and features, we demonstrate two different applications:
from www.cellsignal.com
Haddock is a method for modeling biomolecular complexes using ambiguous interaction restraints and other experimental data. We use anarci [dunbar, j. Here we provide the code to run the antibody protocol of haddock by using the residues belonging to the hypervariable (hv) loops. We find that haddock can generate accurate models of antibodyantigen complexes using an ensemble of antibody. To present the web server’s various interfaces and features, we demonstrate two different applications:
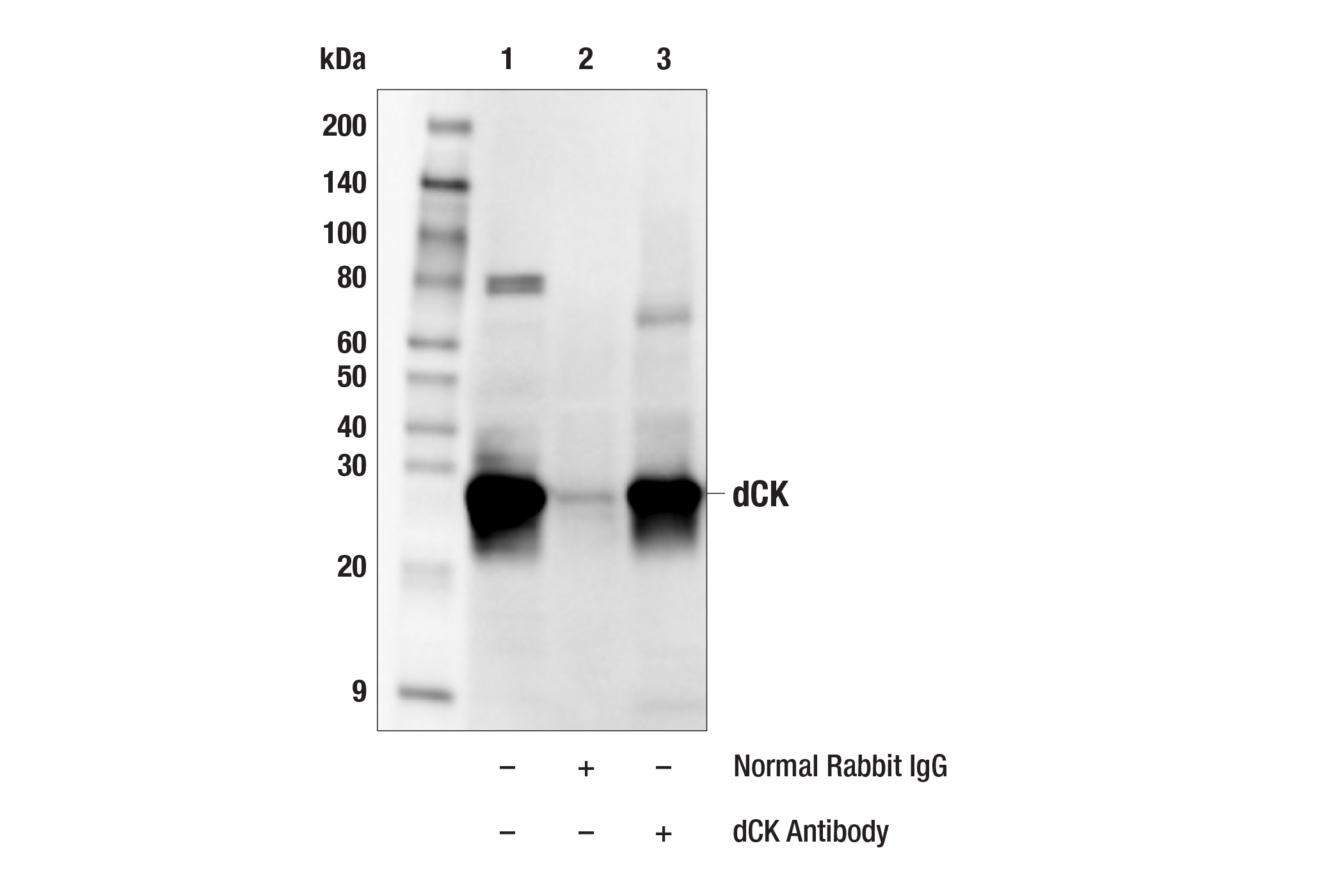
dCK Antibody Cell Signaling Technology
Haddock Antibody Haddock is a method for modeling biomolecular complexes using ambiguous interaction restraints and other experimental data. We find that haddock can generate accurate models of antibodyantigen complexes using an ensemble of antibody. Haddock is a method for modeling biomolecular complexes using ambiguous interaction restraints and other experimental data. To present the web server’s various interfaces and features, we demonstrate two different applications: Here we provide the code to run the antibody protocol of haddock by using the residues belonging to the hypervariable (hv) loops. We use anarci [dunbar, j.
From slideplayer.com
Gydo C.P. van Zundert, Adrien S.J. Melquiond, Alexandre M.J.J. Bonvin ppt download Haddock Antibody We use anarci [dunbar, j. To present the web server’s various interfaces and features, we demonstrate two different applications: Here we provide the code to run the antibody protocol of haddock by using the residues belonging to the hypervariable (hv) loops. Haddock is a method for modeling biomolecular complexes using ambiguous interaction restraints and other experimental data. We find that. Haddock Antibody.
From www.creative-biolabs.com
Multispecific Antibodies Enhance the Effectiveness of Cancer Immunotherapy Creative Biolabs Haddock Antibody We find that haddock can generate accurate models of antibodyantigen complexes using an ensemble of antibody. We use anarci [dunbar, j. Here we provide the code to run the antibody protocol of haddock by using the residues belonging to the hypervariable (hv) loops. To present the web server’s various interfaces and features, we demonstrate two different applications: Haddock is a. Haddock Antibody.
From www.bonvinlab.org
HADDOCK2.4 basic Antibody Antigen tutorial including a comparison with AlphaFold Bonvin Lab Haddock Antibody To present the web server’s various interfaces and features, we demonstrate two different applications: We find that haddock can generate accurate models of antibodyantigen complexes using an ensemble of antibody. Here we provide the code to run the antibody protocol of haddock by using the residues belonging to the hypervariable (hv) loops. Haddock is a method for modeling biomolecular complexes. Haddock Antibody.
From www.thermofisher.cn
IgD Monoclonal Antibody (IA62), eFluor™ 506 (69986842) Haddock Antibody To present the web server’s various interfaces and features, we demonstrate two different applications: Here we provide the code to run the antibody protocol of haddock by using the residues belonging to the hypervariable (hv) loops. We use anarci [dunbar, j. Haddock is a method for modeling biomolecular complexes using ambiguous interaction restraints and other experimental data. We find that. Haddock Antibody.
From www.bonvinlab.org
HADDOCK2.4 basic Antibody Antigen tutorial including a comparison with AlphaFold Bonvin Lab Haddock Antibody Haddock is a method for modeling biomolecular complexes using ambiguous interaction restraints and other experimental data. We use anarci [dunbar, j. We find that haddock can generate accurate models of antibodyantigen complexes using an ensemble of antibody. Here we provide the code to run the antibody protocol of haddock by using the residues belonging to the hypervariable (hv) loops. To. Haddock Antibody.
From www.bonvinlab.org
HADDOCK2.4 basic Antibody Antigen tutorial including a comparison with AlphaFold Bonvin Lab Haddock Antibody To present the web server’s various interfaces and features, we demonstrate two different applications: Haddock is a method for modeling biomolecular complexes using ambiguous interaction restraints and other experimental data. We find that haddock can generate accurate models of antibodyantigen complexes using an ensemble of antibody. We use anarci [dunbar, j. Here we provide the code to run the antibody. Haddock Antibody.
From www.bonvinlab.org
Lowsampling antibodyantigen modelling tutorial using a local version of HADDOCK3 Bonvin Lab Haddock Antibody We use anarci [dunbar, j. Haddock is a method for modeling biomolecular complexes using ambiguous interaction restraints and other experimental data. We find that haddock can generate accurate models of antibodyantigen complexes using an ensemble of antibody. Here we provide the code to run the antibody protocol of haddock by using the residues belonging to the hypervariable (hv) loops. To. Haddock Antibody.
From www.bonvinlab.org
HADDOCK2.4 Antibody Antigen tutorial using PDBtools webserver Bonvin Lab Haddock Antibody We use anarci [dunbar, j. We find that haddock can generate accurate models of antibodyantigen complexes using an ensemble of antibody. Here we provide the code to run the antibody protocol of haddock by using the residues belonging to the hypervariable (hv) loops. Haddock is a method for modeling biomolecular complexes using ambiguous interaction restraints and other experimental data. To. Haddock Antibody.
From www.bonvinlab.org
Lowsampling antibodyantigen modelling tutorial using a local version of HADDOCK3 Bonvin Lab Haddock Antibody Here we provide the code to run the antibody protocol of haddock by using the residues belonging to the hypervariable (hv) loops. To present the web server’s various interfaces and features, we demonstrate two different applications: We use anarci [dunbar, j. Haddock is a method for modeling biomolecular complexes using ambiguous interaction restraints and other experimental data. We find that. Haddock Antibody.
From www.bonvinlab.org
HADDOCK2.4 Antibody Antigen tutorial using PDBtools webserver Bonvin Lab Haddock Antibody We find that haddock can generate accurate models of antibodyantigen complexes using an ensemble of antibody. To present the web server’s various interfaces and features, we demonstrate two different applications: We use anarci [dunbar, j. Here we provide the code to run the antibody protocol of haddock by using the residues belonging to the hypervariable (hv) loops. Haddock is a. Haddock Antibody.
From github.com
GitHub haddocking/HADDOCKantibodyantigen The repository contains material to run the Haddock Antibody Haddock is a method for modeling biomolecular complexes using ambiguous interaction restraints and other experimental data. Here we provide the code to run the antibody protocol of haddock by using the residues belonging to the hypervariable (hv) loops. To present the web server’s various interfaces and features, we demonstrate two different applications: We find that haddock can generate accurate models. Haddock Antibody.
From www.bonvinlab.org
Lowsampling antibodyantigen modelling tutorial using a local version of HADDOCK3 Bonvin Lab Haddock Antibody Here we provide the code to run the antibody protocol of haddock by using the residues belonging to the hypervariable (hv) loops. We use anarci [dunbar, j. To present the web server’s various interfaces and features, we demonstrate two different applications: We find that haddock can generate accurate models of antibodyantigen complexes using an ensemble of antibody. Haddock is a. Haddock Antibody.
From www.researchgate.net
Identification of the connecdennbinding site on AP2. A, Soluble brain... Download Scientific Haddock Antibody Here we provide the code to run the antibody protocol of haddock by using the residues belonging to the hypervariable (hv) loops. We use anarci [dunbar, j. We find that haddock can generate accurate models of antibodyantigen complexes using an ensemble of antibody. To present the web server’s various interfaces and features, we demonstrate two different applications: Haddock is a. Haddock Antibody.
From www.bonvinlab.org
Lowsampling antibodyantigen modelling tutorial using a local version of HADDOCK3 Bonvin Lab Haddock Antibody Here we provide the code to run the antibody protocol of haddock by using the residues belonging to the hypervariable (hv) loops. We use anarci [dunbar, j. Haddock is a method for modeling biomolecular complexes using ambiguous interaction restraints and other experimental data. We find that haddock can generate accurate models of antibodyantigen complexes using an ensemble of antibody. To. Haddock Antibody.
From www.bonvinlab.org
HADDOCK2.4 Antibody Antigen tutorial using PDBtools webserver Bonvin Lab Haddock Antibody Here we provide the code to run the antibody protocol of haddock by using the residues belonging to the hypervariable (hv) loops. We find that haddock can generate accurate models of antibodyantigen complexes using an ensemble of antibody. To present the web server’s various interfaces and features, we demonstrate two different applications: We use anarci [dunbar, j. Haddock is a. Haddock Antibody.
From shop.perensis.com
Haddock Haddock Antibody We find that haddock can generate accurate models of antibodyantigen complexes using an ensemble of antibody. Here we provide the code to run the antibody protocol of haddock by using the residues belonging to the hypervariable (hv) loops. We use anarci [dunbar, j. Haddock is a method for modeling biomolecular complexes using ambiguous interaction restraints and other experimental data. To. Haddock Antibody.
From www.cellsignal.com
AP4B1 Antibody Cell Signaling Technology Haddock Antibody We use anarci [dunbar, j. We find that haddock can generate accurate models of antibodyantigen complexes using an ensemble of antibody. To present the web server’s various interfaces and features, we demonstrate two different applications: Haddock is a method for modeling biomolecular complexes using ambiguous interaction restraints and other experimental data. Here we provide the code to run the antibody. Haddock Antibody.
From www.researchgate.net
Epitope mapping of C2_2E12. (A) Immunoblot analysis of 14 GSTIL33... Download Scientific Diagram Haddock Antibody Haddock is a method for modeling biomolecular complexes using ambiguous interaction restraints and other experimental data. To present the web server’s various interfaces and features, we demonstrate two different applications: We find that haddock can generate accurate models of antibodyantigen complexes using an ensemble of antibody. We use anarci [dunbar, j. Here we provide the code to run the antibody. Haddock Antibody.
From www.bonvinlab.org
HADDOCK2.4 basic Antibody Antigen tutorial including a comparison with AlphaFold Bonvin Lab Haddock Antibody We use anarci [dunbar, j. Haddock is a method for modeling biomolecular complexes using ambiguous interaction restraints and other experimental data. We find that haddock can generate accurate models of antibodyantigen complexes using an ensemble of antibody. To present the web server’s various interfaces and features, we demonstrate two different applications: Here we provide the code to run the antibody. Haddock Antibody.
From www.bonvinlab.org
Lowsampling antibodyantigen modelling tutorial using a local version of HADDOCK3 Bonvin Lab Haddock Antibody Here we provide the code to run the antibody protocol of haddock by using the residues belonging to the hypervariable (hv) loops. Haddock is a method for modeling biomolecular complexes using ambiguous interaction restraints and other experimental data. We use anarci [dunbar, j. We find that haddock can generate accurate models of antibodyantigen complexes using an ensemble of antibody. To. Haddock Antibody.
From www.bonvinlab.org
Lowsampling antibodyantigen modelling tutorial using a local version of HADDOCK3 Bonvin Lab Haddock Antibody We use anarci [dunbar, j. We find that haddock can generate accurate models of antibodyantigen complexes using an ensemble of antibody. Haddock is a method for modeling biomolecular complexes using ambiguous interaction restraints and other experimental data. Here we provide the code to run the antibody protocol of haddock by using the residues belonging to the hypervariable (hv) loops. To. Haddock Antibody.
From www.researchgate.net
A model of the complete IgNAR antibody. (A) A structural model of... Download Scientific Diagram Haddock Antibody We find that haddock can generate accurate models of antibodyantigen complexes using an ensemble of antibody. Here we provide the code to run the antibody protocol of haddock by using the residues belonging to the hypervariable (hv) loops. We use anarci [dunbar, j. Haddock is a method for modeling biomolecular complexes using ambiguous interaction restraints and other experimental data. To. Haddock Antibody.
From bioexcel.eu
Software Bioexcel Haddock Antibody Here we provide the code to run the antibody protocol of haddock by using the residues belonging to the hypervariable (hv) loops. We find that haddock can generate accurate models of antibodyantigen complexes using an ensemble of antibody. We use anarci [dunbar, j. To present the web server’s various interfaces and features, we demonstrate two different applications: Haddock is a. Haddock Antibody.
From nmk.world
Haddock vs Pollock The Key Differences Haddock Antibody Here we provide the code to run the antibody protocol of haddock by using the residues belonging to the hypervariable (hv) loops. We use anarci [dunbar, j. To present the web server’s various interfaces and features, we demonstrate two different applications: We find that haddock can generate accurate models of antibodyantigen complexes using an ensemble of antibody. Haddock is a. Haddock Antibody.
From www.bonvinlab.org
HADDOCK2.4 basic Antibody Antigen tutorial including a comparison with AlphaFold Bonvin Lab Haddock Antibody We use anarci [dunbar, j. We find that haddock can generate accurate models of antibodyantigen complexes using an ensemble of antibody. Haddock is a method for modeling biomolecular complexes using ambiguous interaction restraints and other experimental data. Here we provide the code to run the antibody protocol of haddock by using the residues belonging to the hypervariable (hv) loops. To. Haddock Antibody.
From www.researchgate.net
Computational analysis of p95HER2 binding to Oslo2 mAb. (a) A heatmap... Download Scientific Haddock Antibody Here we provide the code to run the antibody protocol of haddock by using the residues belonging to the hypervariable (hv) loops. We find that haddock can generate accurate models of antibodyantigen complexes using an ensemble of antibody. Haddock is a method for modeling biomolecular complexes using ambiguous interaction restraints and other experimental data. We use anarci [dunbar, j. To. Haddock Antibody.
From pipebio.com
Machine learning in antibody discovery and engineering Haddock Antibody Here we provide the code to run the antibody protocol of haddock by using the residues belonging to the hypervariable (hv) loops. We use anarci [dunbar, j. To present the web server’s various interfaces and features, we demonstrate two different applications: Haddock is a method for modeling biomolecular complexes using ambiguous interaction restraints and other experimental data. We find that. Haddock Antibody.
From www.bonvinlab.org
Antibodyantigen modelling tutorial using a local version of HADDOCK3 Bonvin Lab Haddock Antibody Haddock is a method for modeling biomolecular complexes using ambiguous interaction restraints and other experimental data. We find that haddock can generate accurate models of antibodyantigen complexes using an ensemble of antibody. To present the web server’s various interfaces and features, we demonstrate two different applications: We use anarci [dunbar, j. Here we provide the code to run the antibody. Haddock Antibody.
From www.researchgate.net
(PDF) A protocol for informationdriven antibodyantigen modelling with the HADDOCK2.4 webserver Haddock Antibody We use anarci [dunbar, j. We find that haddock can generate accurate models of antibodyantigen complexes using an ensemble of antibody. To present the web server’s various interfaces and features, we demonstrate two different applications: Here we provide the code to run the antibody protocol of haddock by using the residues belonging to the hypervariable (hv) loops. Haddock is a. Haddock Antibody.
From www.bonvinlab.org
Lowsampling antibodyantigen modelling tutorial using a local version of HADDOCK3 Bonvin Lab Haddock Antibody Haddock is a method for modeling biomolecular complexes using ambiguous interaction restraints and other experimental data. To present the web server’s various interfaces and features, we demonstrate two different applications: Here we provide the code to run the antibody protocol of haddock by using the residues belonging to the hypervariable (hv) loops. We use anarci [dunbar, j. We find that. Haddock Antibody.
From www.bonvinlab.org
Antibodyantigen modelling tutorial using a local version of HADDOCK3 Bonvin Lab Haddock Antibody We find that haddock can generate accurate models of antibodyantigen complexes using an ensemble of antibody. Here we provide the code to run the antibody protocol of haddock by using the residues belonging to the hypervariable (hv) loops. We use anarci [dunbar, j. To present the web server’s various interfaces and features, we demonstrate two different applications: Haddock is a. Haddock Antibody.
From www.researchgate.net
Sergey OVCHINNIKOV Harvard University, MA Harvard FAS Center for Systems Biology Haddock Antibody To present the web server’s various interfaces and features, we demonstrate two different applications: We use anarci [dunbar, j. We find that haddock can generate accurate models of antibodyantigen complexes using an ensemble of antibody. Haddock is a method for modeling biomolecular complexes using ambiguous interaction restraints and other experimental data. Here we provide the code to run the antibody. Haddock Antibody.
From www.cellsignal.com
dCK Antibody Cell Signaling Technology Haddock Antibody We find that haddock can generate accurate models of antibodyantigen complexes using an ensemble of antibody. Here we provide the code to run the antibody protocol of haddock by using the residues belonging to the hypervariable (hv) loops. To present the web server’s various interfaces and features, we demonstrate two different applications: We use anarci [dunbar, j. Haddock is a. Haddock Antibody.
From www.researchgate.net
Simulated docking of antibody 60H07 with the Ravn GP antigen. The... Download Scientific Diagram Haddock Antibody To present the web server’s various interfaces and features, we demonstrate two different applications: We use anarci [dunbar, j. Here we provide the code to run the antibody protocol of haddock by using the residues belonging to the hypervariable (hv) loops. We find that haddock can generate accurate models of antibodyantigen complexes using an ensemble of antibody. Haddock is a. Haddock Antibody.
From www.researchgate.net
CD8 stalk sequences and modeling of a novel CD8abpMHC binding mode.... Download Scientific Haddock Antibody Haddock is a method for modeling biomolecular complexes using ambiguous interaction restraints and other experimental data. Here we provide the code to run the antibody protocol of haddock by using the residues belonging to the hypervariable (hv) loops. We find that haddock can generate accurate models of antibodyantigen complexes using an ensemble of antibody. We use anarci [dunbar, j. To. Haddock Antibody.