Differential Gene Expression Glm . To test for de genes between two specific groups of. In contrast to exact tests, glms allow for. In this tutorial, we will give you an overview of the deseq2 pipeline to find differentially expressed genes between two conditions. Deseq2 is an r/bioconductor implemented method to detect. A statistical test based on the negative binomial distribution (via a generalized linear model, glm) can be used to assess differential expression for each gene.
from cbirt.net
A statistical test based on the negative binomial distribution (via a generalized linear model, glm) can be used to assess differential expression for each gene. In contrast to exact tests, glms allow for. In this tutorial, we will give you an overview of the deseq2 pipeline to find differentially expressed genes between two conditions. To test for de genes between two specific groups of. Deseq2 is an r/bioconductor implemented method to detect.
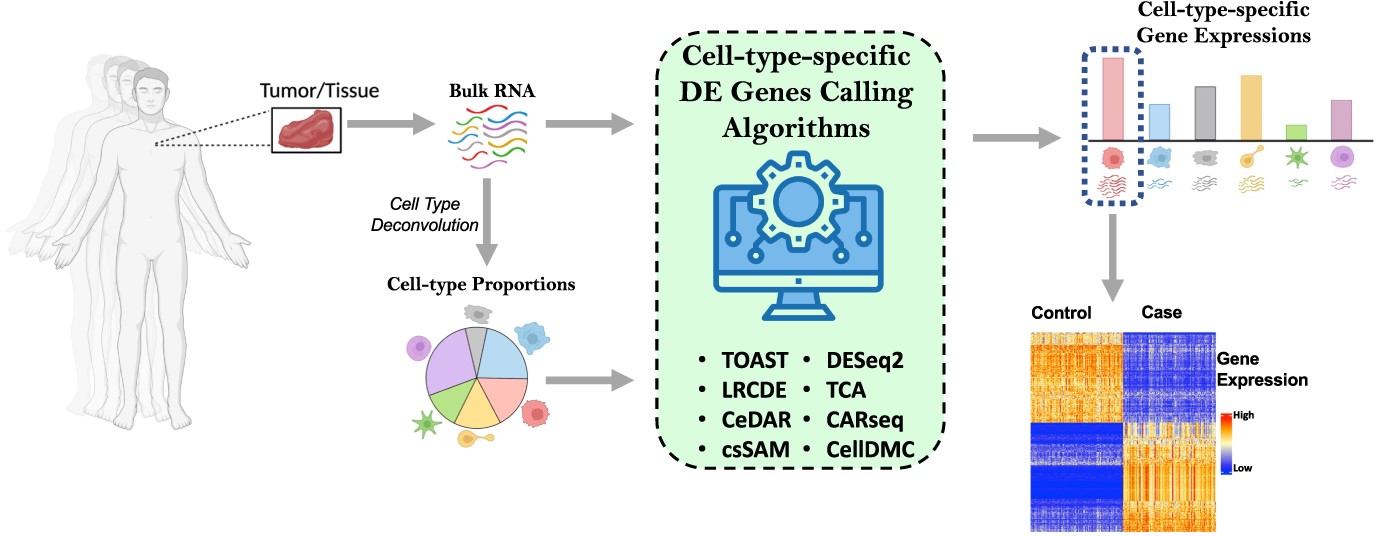
Cell TypeSpecific Differential Gene Expression A Guide Through The
Differential Gene Expression Glm In this tutorial, we will give you an overview of the deseq2 pipeline to find differentially expressed genes between two conditions. In this tutorial, we will give you an overview of the deseq2 pipeline to find differentially expressed genes between two conditions. Deseq2 is an r/bioconductor implemented method to detect. A statistical test based on the negative binomial distribution (via a generalized linear model, glm) can be used to assess differential expression for each gene. To test for de genes between two specific groups of. In contrast to exact tests, glms allow for.
From www.researchgate.net
Differential gene expression in serial isolates. (A) Hierarchical Differential Gene Expression Glm Deseq2 is an r/bioconductor implemented method to detect. To test for de genes between two specific groups of. In contrast to exact tests, glms allow for. In this tutorial, we will give you an overview of the deseq2 pipeline to find differentially expressed genes between two conditions. A statistical test based on the negative binomial distribution (via a generalized linear. Differential Gene Expression Glm.
From www.pnas.org
Predictability of human differential gene expression PNAS Differential Gene Expression Glm To test for de genes between two specific groups of. In contrast to exact tests, glms allow for. In this tutorial, we will give you an overview of the deseq2 pipeline to find differentially expressed genes between two conditions. A statistical test based on the negative binomial distribution (via a generalized linear model, glm) can be used to assess differential. Differential Gene Expression Glm.
From www.researchgate.net
Differential gene expression and pathway analysis in response to Differential Gene Expression Glm Deseq2 is an r/bioconductor implemented method to detect. To test for de genes between two specific groups of. In this tutorial, we will give you an overview of the deseq2 pipeline to find differentially expressed genes between two conditions. In contrast to exact tests, glms allow for. A statistical test based on the negative binomial distribution (via a generalized linear. Differential Gene Expression Glm.
From www.researchgate.net
Differential expression analysis of IL8 expression in myeloid cells a Differential Gene Expression Glm In contrast to exact tests, glms allow for. Deseq2 is an r/bioconductor implemented method to detect. A statistical test based on the negative binomial distribution (via a generalized linear model, glm) can be used to assess differential expression for each gene. To test for de genes between two specific groups of. In this tutorial, we will give you an overview. Differential Gene Expression Glm.
From www.researchgate.net
Differential gene expression analysis. a Venn diagram of gene Differential Gene Expression Glm Deseq2 is an r/bioconductor implemented method to detect. A statistical test based on the negative binomial distribution (via a generalized linear model, glm) can be used to assess differential expression for each gene. In this tutorial, we will give you an overview of the deseq2 pipeline to find differentially expressed genes between two conditions. To test for de genes between. Differential Gene Expression Glm.
From www.researchgate.net
WGCNA of differential gene expressions and the related traits. (A Differential Gene Expression Glm In this tutorial, we will give you an overview of the deseq2 pipeline to find differentially expressed genes between two conditions. To test for de genes between two specific groups of. In contrast to exact tests, glms allow for. A statistical test based on the negative binomial distribution (via a generalized linear model, glm) can be used to assess differential. Differential Gene Expression Glm.
From www.researchgate.net
Overall gene expression and differential expression analysis. (A Differential Gene Expression Glm A statistical test based on the negative binomial distribution (via a generalized linear model, glm) can be used to assess differential expression for each gene. Deseq2 is an r/bioconductor implemented method to detect. In this tutorial, we will give you an overview of the deseq2 pipeline to find differentially expressed genes between two conditions. In contrast to exact tests, glms. Differential Gene Expression Glm.
From www.researchgate.net
Differential gene expression analysis comparing gene expression levels Differential Gene Expression Glm To test for de genes between two specific groups of. In contrast to exact tests, glms allow for. In this tutorial, we will give you an overview of the deseq2 pipeline to find differentially expressed genes between two conditions. Deseq2 is an r/bioconductor implemented method to detect. A statistical test based on the negative binomial distribution (via a generalized linear. Differential Gene Expression Glm.
From www.researchgate.net
Differential geneexpression analysis and enriched gene ontology Differential Gene Expression Glm To test for de genes between two specific groups of. In contrast to exact tests, glms allow for. A statistical test based on the negative binomial distribution (via a generalized linear model, glm) can be used to assess differential expression for each gene. Deseq2 is an r/bioconductor implemented method to detect. In this tutorial, we will give you an overview. Differential Gene Expression Glm.
From www.researchgate.net
Differential gene expression. (A) Principal component analysis (PCA Differential Gene Expression Glm A statistical test based on the negative binomial distribution (via a generalized linear model, glm) can be used to assess differential expression for each gene. In this tutorial, we will give you an overview of the deseq2 pipeline to find differentially expressed genes between two conditions. In contrast to exact tests, glms allow for. Deseq2 is an r/bioconductor implemented method. Differential Gene Expression Glm.
From www.sc-best-practices.org
16. Differential gene expression analysis — Singlecell best practices Differential Gene Expression Glm In contrast to exact tests, glms allow for. To test for de genes between two specific groups of. In this tutorial, we will give you an overview of the deseq2 pipeline to find differentially expressed genes between two conditions. Deseq2 is an r/bioconductor implemented method to detect. A statistical test based on the negative binomial distribution (via a generalized linear. Differential Gene Expression Glm.
From www.researchgate.net
Differential gene expression analysis results, including principle Differential Gene Expression Glm In this tutorial, we will give you an overview of the deseq2 pipeline to find differentially expressed genes between two conditions. In contrast to exact tests, glms allow for. Deseq2 is an r/bioconductor implemented method to detect. To test for de genes between two specific groups of. A statistical test based on the negative binomial distribution (via a generalized linear. Differential Gene Expression Glm.
From www.researchgate.net
Interaction plots for pairs of explanatory variables in overall GLM Differential Gene Expression Glm In contrast to exact tests, glms allow for. In this tutorial, we will give you an overview of the deseq2 pipeline to find differentially expressed genes between two conditions. A statistical test based on the negative binomial distribution (via a generalized linear model, glm) can be used to assess differential expression for each gene. Deseq2 is an r/bioconductor implemented method. Differential Gene Expression Glm.
From www.researchgate.net
Differential gene expression by 1,25(OH) 2 D or hnRNPC1/2 knockdown Differential Gene Expression Glm In this tutorial, we will give you an overview of the deseq2 pipeline to find differentially expressed genes between two conditions. A statistical test based on the negative binomial distribution (via a generalized linear model, glm) can be used to assess differential expression for each gene. In contrast to exact tests, glms allow for. To test for de genes between. Differential Gene Expression Glm.
From www.researchgate.net
Differential gene expression analysis using DESeq and GLM analysis to Differential Gene Expression Glm To test for de genes between two specific groups of. A statistical test based on the negative binomial distribution (via a generalized linear model, glm) can be used to assess differential expression for each gene. In contrast to exact tests, glms allow for. In this tutorial, we will give you an overview of the deseq2 pipeline to find differentially expressed. Differential Gene Expression Glm.
From cbirt.net
Cell TypeSpecific Differential Gene Expression A Guide Through The Differential Gene Expression Glm Deseq2 is an r/bioconductor implemented method to detect. A statistical test based on the negative binomial distribution (via a generalized linear model, glm) can be used to assess differential expression for each gene. In contrast to exact tests, glms allow for. To test for de genes between two specific groups of. In this tutorial, we will give you an overview. Differential Gene Expression Glm.
From www.researchgate.net
Differential gene expression and pathway enrichment analysis for Differential Gene Expression Glm To test for de genes between two specific groups of. Deseq2 is an r/bioconductor implemented method to detect. A statistical test based on the negative binomial distribution (via a generalized linear model, glm) can be used to assess differential expression for each gene. In contrast to exact tests, glms allow for. In this tutorial, we will give you an overview. Differential Gene Expression Glm.
From www.researchgate.net
Differential expression analysis of response in myeloid cells a Differential Gene Expression Glm Deseq2 is an r/bioconductor implemented method to detect. In contrast to exact tests, glms allow for. A statistical test based on the negative binomial distribution (via a generalized linear model, glm) can be used to assess differential expression for each gene. In this tutorial, we will give you an overview of the deseq2 pipeline to find differentially expressed genes between. Differential Gene Expression Glm.
From www.researchgate.net
Differential gene expression analysis and visualisation options. (A Differential Gene Expression Glm Deseq2 is an r/bioconductor implemented method to detect. To test for de genes between two specific groups of. In this tutorial, we will give you an overview of the deseq2 pipeline to find differentially expressed genes between two conditions. A statistical test based on the negative binomial distribution (via a generalized linear model, glm) can be used to assess differential. Differential Gene Expression Glm.
From www.researchgate.net
Differential gene expression following positive or negative selection Differential Gene Expression Glm Deseq2 is an r/bioconductor implemented method to detect. In contrast to exact tests, glms allow for. A statistical test based on the negative binomial distribution (via a generalized linear model, glm) can be used to assess differential expression for each gene. In this tutorial, we will give you an overview of the deseq2 pipeline to find differentially expressed genes between. Differential Gene Expression Glm.
From www.researchgate.net
Differential gene expression analysis between BluePrint single and dual Differential Gene Expression Glm A statistical test based on the negative binomial distribution (via a generalized linear model, glm) can be used to assess differential expression for each gene. In contrast to exact tests, glms allow for. Deseq2 is an r/bioconductor implemented method to detect. In this tutorial, we will give you an overview of the deseq2 pipeline to find differentially expressed genes between. Differential Gene Expression Glm.
From www.slideserve.com
PPT Differential Gene Expression PowerPoint Presentation, free Differential Gene Expression Glm In contrast to exact tests, glms allow for. Deseq2 is an r/bioconductor implemented method to detect. To test for de genes between two specific groups of. A statistical test based on the negative binomial distribution (via a generalized linear model, glm) can be used to assess differential expression for each gene. In this tutorial, we will give you an overview. Differential Gene Expression Glm.
From www.researchgate.net
Differential gene expression profiles of overlapping and unique Differential Gene Expression Glm In contrast to exact tests, glms allow for. A statistical test based on the negative binomial distribution (via a generalized linear model, glm) can be used to assess differential expression for each gene. Deseq2 is an r/bioconductor implemented method to detect. In this tutorial, we will give you an overview of the deseq2 pipeline to find differentially expressed genes between. Differential Gene Expression Glm.
From www.researchgate.net
of differential gene expression analysis patterns. (A) Venn diagrams Differential Gene Expression Glm A statistical test based on the negative binomial distribution (via a generalized linear model, glm) can be used to assess differential expression for each gene. To test for de genes between two specific groups of. Deseq2 is an r/bioconductor implemented method to detect. In contrast to exact tests, glms allow for. In this tutorial, we will give you an overview. Differential Gene Expression Glm.
From www.researchgate.net
AR regulation is antagonized by ETV1 overexpression. a The generalized Differential Gene Expression Glm In this tutorial, we will give you an overview of the deseq2 pipeline to find differentially expressed genes between two conditions. Deseq2 is an r/bioconductor implemented method to detect. In contrast to exact tests, glms allow for. A statistical test based on the negative binomial distribution (via a generalized linear model, glm) can be used to assess differential expression for. Differential Gene Expression Glm.
From www.researchgate.net
Differential gene expression. a Principal component analysis of the Differential Gene Expression Glm A statistical test based on the negative binomial distribution (via a generalized linear model, glm) can be used to assess differential expression for each gene. To test for de genes between two specific groups of. In contrast to exact tests, glms allow for. Deseq2 is an r/bioconductor implemented method to detect. In this tutorial, we will give you an overview. Differential Gene Expression Glm.
From www.researchgate.net
A. Clustering diagram of differential gene expression patterns among Differential Gene Expression Glm To test for de genes between two specific groups of. In this tutorial, we will give you an overview of the deseq2 pipeline to find differentially expressed genes between two conditions. A statistical test based on the negative binomial distribution (via a generalized linear model, glm) can be used to assess differential expression for each gene. In contrast to exact. Differential Gene Expression Glm.
From www.researchgate.net
Differential gene expression profiles of Anemonia viridis and Differential Gene Expression Glm In this tutorial, we will give you an overview of the deseq2 pipeline to find differentially expressed genes between two conditions. Deseq2 is an r/bioconductor implemented method to detect. In contrast to exact tests, glms allow for. A statistical test based on the negative binomial distribution (via a generalized linear model, glm) can be used to assess differential expression for. Differential Gene Expression Glm.
From www.researchgate.net
Differential gene expression analysis in A. suecica Patterns of Differential Gene Expression Glm In this tutorial, we will give you an overview of the deseq2 pipeline to find differentially expressed genes between two conditions. A statistical test based on the negative binomial distribution (via a generalized linear model, glm) can be used to assess differential expression for each gene. Deseq2 is an r/bioconductor implemented method to detect. In contrast to exact tests, glms. Differential Gene Expression Glm.
From www.researchgate.net
Differential gene expression analysis and enrichment analysis. (A Differential Gene Expression Glm To test for de genes between two specific groups of. A statistical test based on the negative binomial distribution (via a generalized linear model, glm) can be used to assess differential expression for each gene. In contrast to exact tests, glms allow for. In this tutorial, we will give you an overview of the deseq2 pipeline to find differentially expressed. Differential Gene Expression Glm.
From www.researchgate.net
Differential gene expression analysis for MAIT cells from patients and Differential Gene Expression Glm Deseq2 is an r/bioconductor implemented method to detect. To test for de genes between two specific groups of. In this tutorial, we will give you an overview of the deseq2 pipeline to find differentially expressed genes between two conditions. In contrast to exact tests, glms allow for. A statistical test based on the negative binomial distribution (via a generalized linear. Differential Gene Expression Glm.
From www.slideserve.com
PPT CHAPTER 18 PowerPoint Presentation ID5719999 Differential Gene Expression Glm In contrast to exact tests, glms allow for. In this tutorial, we will give you an overview of the deseq2 pipeline to find differentially expressed genes between two conditions. Deseq2 is an r/bioconductor implemented method to detect. To test for de genes between two specific groups of. A statistical test based on the negative binomial distribution (via a generalized linear. Differential Gene Expression Glm.
From www.researchgate.net
Differential gene expression profile. (a) Principal component analysis Differential Gene Expression Glm In this tutorial, we will give you an overview of the deseq2 pipeline to find differentially expressed genes between two conditions. To test for de genes between two specific groups of. In contrast to exact tests, glms allow for. Deseq2 is an r/bioconductor implemented method to detect. A statistical test based on the negative binomial distribution (via a generalized linear. Differential Gene Expression Glm.
From www.researchgate.net
(A) Differential expression of ATP7A gene in scurred and SHE groups Differential Gene Expression Glm In this tutorial, we will give you an overview of the deseq2 pipeline to find differentially expressed genes between two conditions. A statistical test based on the negative binomial distribution (via a generalized linear model, glm) can be used to assess differential expression for each gene. Deseq2 is an r/bioconductor implemented method to detect. To test for de genes between. Differential Gene Expression Glm.
From www.researchgate.net
Differential gene expression and celltype composition a,b Differential Gene Expression Glm To test for de genes between two specific groups of. In this tutorial, we will give you an overview of the deseq2 pipeline to find differentially expressed genes between two conditions. Deseq2 is an r/bioconductor implemented method to detect. In contrast to exact tests, glms allow for. A statistical test based on the negative binomial distribution (via a generalized linear. Differential Gene Expression Glm.