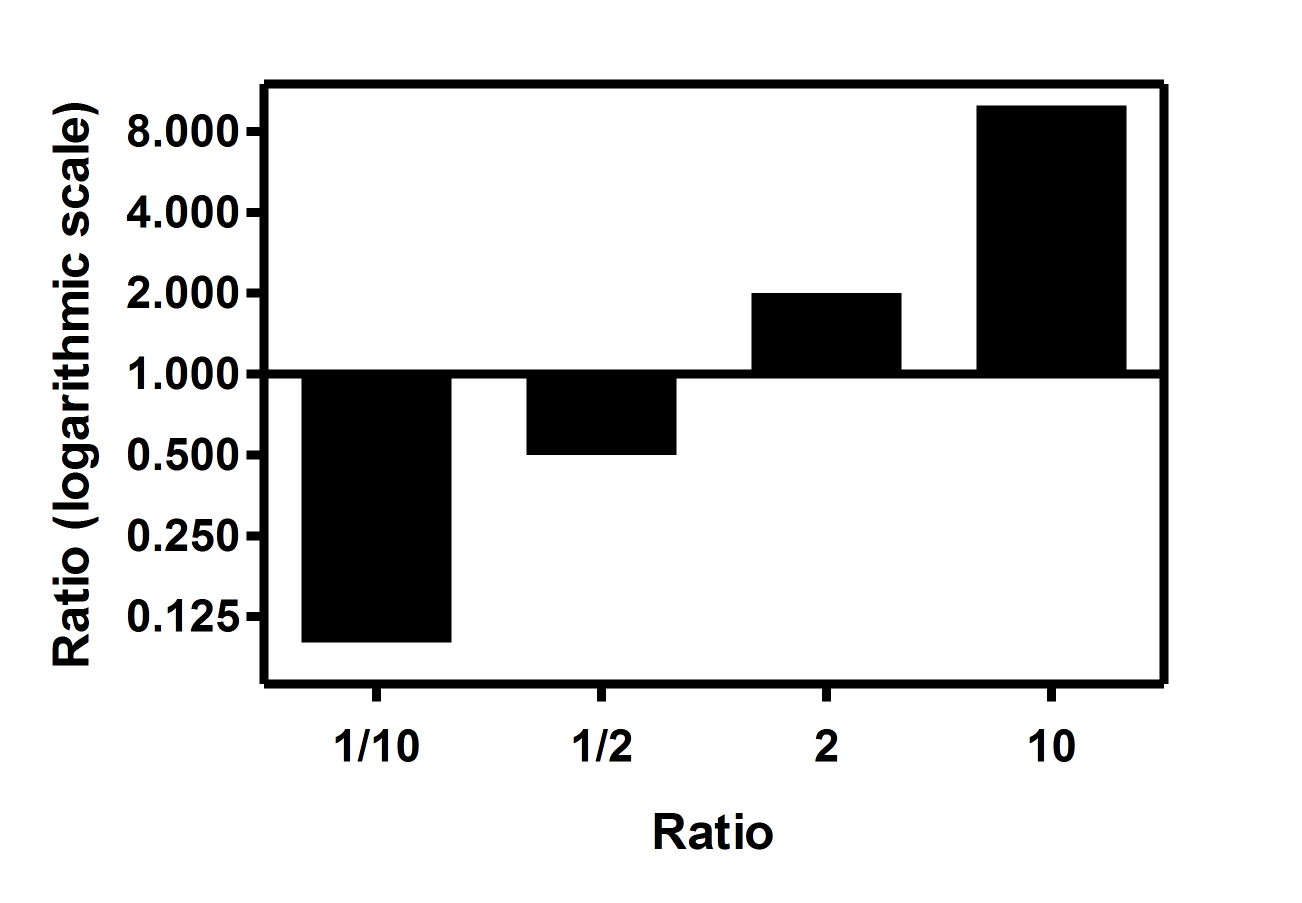
Logarithmic Fold Change . We propose a fold change transform that demonstrates a combination of visualization properties exhibited by log and linear plots of fold. Log2 fold change values (eg 1 or 2 or 3) can be converted to fold changes by taking 2^1 or 2^2 or 2^3 = 1 or 4 or 8; In gene expression analysis, such as microarray or rna. Log fold change is a statistical measure used to describe the magnitude of change in expression levels of genes between two different conditions. The most common approach in the comparative analysis of transcriptomics data is to test the null hypothesis that the logarithmic fold. The log2 fold change (log2fc) term is commonly used in bioinformatics to measure the changes in gene expression between two conditions (e.g. Log2 fold change is the ratio of expression levels between two conditions and it is calculated by taking log2 of the ratio of expression levels of two conditions. The log2 (log with base 2) is most commonly used. Fold change is a metric that expresses the extent of change between two measurements.
from exyjvtzky.blob.core.windows.net
Log2 fold change is the ratio of expression levels between two conditions and it is calculated by taking log2 of the ratio of expression levels of two conditions. We propose a fold change transform that demonstrates a combination of visualization properties exhibited by log and linear plots of fold. Fold change is a metric that expresses the extent of change between two measurements. Log fold change is a statistical measure used to describe the magnitude of change in expression levels of genes between two different conditions. In gene expression analysis, such as microarray or rna. Log2 fold change values (eg 1 or 2 or 3) can be converted to fold changes by taking 2^1 or 2^2 or 2^3 = 1 or 4 or 8; The log2 (log with base 2) is most commonly used. The most common approach in the comparative analysis of transcriptomics data is to test the null hypothesis that the logarithmic fold. The log2 fold change (log2fc) term is commonly used in bioinformatics to measure the changes in gene expression between two conditions (e.g.
Graphpad Log Scale at Christopher Roman blog
Logarithmic Fold Change The most common approach in the comparative analysis of transcriptomics data is to test the null hypothesis that the logarithmic fold. Log2 fold change is the ratio of expression levels between two conditions and it is calculated by taking log2 of the ratio of expression levels of two conditions. Log fold change is a statistical measure used to describe the magnitude of change in expression levels of genes between two different conditions. We propose a fold change transform that demonstrates a combination of visualization properties exhibited by log and linear plots of fold. The most common approach in the comparative analysis of transcriptomics data is to test the null hypothesis that the logarithmic fold. In gene expression analysis, such as microarray or rna. The log2 (log with base 2) is most commonly used. Fold change is a metric that expresses the extent of change between two measurements. Log2 fold change values (eg 1 or 2 or 3) can be converted to fold changes by taking 2^1 or 2^2 or 2^3 = 1 or 4 or 8; The log2 fold change (log2fc) term is commonly used in bioinformatics to measure the changes in gene expression between two conditions (e.g.
From www.researchgate.net
MDplot showing the logfold change of differential expressed lncRNAs Logarithmic Fold Change The log2 fold change (log2fc) term is commonly used in bioinformatics to measure the changes in gene expression between two conditions (e.g. We propose a fold change transform that demonstrates a combination of visualization properties exhibited by log and linear plots of fold. In gene expression analysis, such as microarray or rna. Log2 fold change is the ratio of expression. Logarithmic Fold Change.
From www.graphpad.com
Graphing data expressed as fold changes, or ratios. FAQ 1721 GraphPad Logarithmic Fold Change The most common approach in the comparative analysis of transcriptomics data is to test the null hypothesis that the logarithmic fold. Log fold change is a statistical measure used to describe the magnitude of change in expression levels of genes between two different conditions. The log2 fold change (log2fc) term is commonly used in bioinformatics to measure the changes in. Logarithmic Fold Change.
From www.researchgate.net
a Log2fold change in normalized counts between the mixture tissue and Logarithmic Fold Change In gene expression analysis, such as microarray or rna. We propose a fold change transform that demonstrates a combination of visualization properties exhibited by log and linear plots of fold. Fold change is a metric that expresses the extent of change between two measurements. Log2 fold change is the ratio of expression levels between two conditions and it is calculated. Logarithmic Fold Change.
From www.biostars.org
Could you please explain Fold change, of change, and log2 fold change Logarithmic Fold Change The log2 fold change (log2fc) term is commonly used in bioinformatics to measure the changes in gene expression between two conditions (e.g. Log2 fold change values (eg 1 or 2 or 3) can be converted to fold changes by taking 2^1 or 2^2 or 2^3 = 1 or 4 or 8; We propose a fold change transform that demonstrates a. Logarithmic Fold Change.
From mathsathome.com
How to Change the Base of a Logarithm Logarithmic Fold Change Log fold change is a statistical measure used to describe the magnitude of change in expression levels of genes between two different conditions. Log2 fold change is the ratio of expression levels between two conditions and it is calculated by taking log2 of the ratio of expression levels of two conditions. The log2 (log with base 2) is most commonly. Logarithmic Fold Change.
From www.researchgate.net
Scatter plot of the logfoldchanges of methylation levels in gene Logarithmic Fold Change The log2 (log with base 2) is most commonly used. Log2 fold change is the ratio of expression levels between two conditions and it is calculated by taking log2 of the ratio of expression levels of two conditions. Log fold change is a statistical measure used to describe the magnitude of change in expression levels of genes between two different. Logarithmic Fold Change.
From exyjvtzky.blob.core.windows.net
Graphpad Log Scale at Christopher Roman blog Logarithmic Fold Change Log2 fold change values (eg 1 or 2 or 3) can be converted to fold changes by taking 2^1 or 2^2 or 2^3 = 1 or 4 or 8; The most common approach in the comparative analysis of transcriptomics data is to test the null hypothesis that the logarithmic fold. Fold change is a metric that expresses the extent of. Logarithmic Fold Change.
From www.researchgate.net
Correlation between the log2(fold change) in gene expression and the Logarithmic Fold Change The log2 (log with base 2) is most commonly used. Log fold change is a statistical measure used to describe the magnitude of change in expression levels of genes between two different conditions. The most common approach in the comparative analysis of transcriptomics data is to test the null hypothesis that the logarithmic fold. Fold change is a metric that. Logarithmic Fold Change.
From www.researchgate.net
Heatmap of log 2 foldchange value of differently expression genes in Logarithmic Fold Change Log2 fold change is the ratio of expression levels between two conditions and it is calculated by taking log2 of the ratio of expression levels of two conditions. The log2 fold change (log2fc) term is commonly used in bioinformatics to measure the changes in gene expression between two conditions (e.g. Log2 fold change values (eg 1 or 2 or 3). Logarithmic Fold Change.
From www.bioz.com
Log2 Fold Changes GraphPad Software Inc Bioz Logarithmic Fold Change Log fold change is a statistical measure used to describe the magnitude of change in expression levels of genes between two different conditions. Fold change is a metric that expresses the extent of change between two measurements. The log2 fold change (log2fc) term is commonly used in bioinformatics to measure the changes in gene expression between two conditions (e.g. We. Logarithmic Fold Change.
From www.researchgate.net
(A) Comparison between the log2(G/E) (log foldchange) estimates Logarithmic Fold Change We propose a fold change transform that demonstrates a combination of visualization properties exhibited by log and linear plots of fold. In gene expression analysis, such as microarray or rna. Fold change is a metric that expresses the extent of change between two measurements. The log2 (log with base 2) is most commonly used. The log2 fold change (log2fc) term. Logarithmic Fold Change.
From www.researchgate.net
The generalized fold change extends the established (medianbased) fold Logarithmic Fold Change The log2 (log with base 2) is most commonly used. Log2 fold change values (eg 1 or 2 or 3) can be converted to fold changes by taking 2^1 or 2^2 or 2^3 = 1 or 4 or 8; We propose a fold change transform that demonstrates a combination of visualization properties exhibited by log and linear plots of fold.. Logarithmic Fold Change.
From www.researchgate.net
Volcano plot showing log2 (fold change, FC) against −log10 (pvalue) of Logarithmic Fold Change The most common approach in the comparative analysis of transcriptomics data is to test the null hypothesis that the logarithmic fold. Log2 fold change is the ratio of expression levels between two conditions and it is calculated by taking log2 of the ratio of expression levels of two conditions. Log2 fold change values (eg 1 or 2 or 3) can. Logarithmic Fold Change.
From www.researchgate.net
Boxplots showing gene expression (fold change (FC), log2scale) in Logarithmic Fold Change Fold change is a metric that expresses the extent of change between two measurements. The log2 (log with base 2) is most commonly used. Log2 fold change values (eg 1 or 2 or 3) can be converted to fold changes by taking 2^1 or 2^2 or 2^3 = 1 or 4 or 8; The log2 fold change (log2fc) term is. Logarithmic Fold Change.
From www.researchgate.net
Log2 transformed Fold Change ΔCq values form qPCR and TPM from RNAseq Logarithmic Fold Change Log fold change is a statistical measure used to describe the magnitude of change in expression levels of genes between two different conditions. We propose a fold change transform that demonstrates a combination of visualization properties exhibited by log and linear plots of fold. Log2 fold change values (eg 1 or 2 or 3) can be converted to fold changes. Logarithmic Fold Change.
From www.researchgate.net
Relative expression (Fold change in logarithmic graphics) analysis of Logarithmic Fold Change We propose a fold change transform that demonstrates a combination of visualization properties exhibited by log and linear plots of fold. Log fold change is a statistical measure used to describe the magnitude of change in expression levels of genes between two different conditions. The log2 (log with base 2) is most commonly used. Log2 fold change is the ratio. Logarithmic Fold Change.
From www.researchgate.net
Heatmap of logfold change (LFC) comparisons of differential gene Logarithmic Fold Change Fold change is a metric that expresses the extent of change between two measurements. In gene expression analysis, such as microarray or rna. The most common approach in the comparative analysis of transcriptomics data is to test the null hypothesis that the logarithmic fold. We propose a fold change transform that demonstrates a combination of visualization properties exhibited by log. Logarithmic Fold Change.
From www.researchgate.net
Volcano plots of microarray data showing the relationship between fold Logarithmic Fold Change The log2 fold change (log2fc) term is commonly used in bioinformatics to measure the changes in gene expression between two conditions (e.g. The most common approach in the comparative analysis of transcriptomics data is to test the null hypothesis that the logarithmic fold. Log2 fold change is the ratio of expression levels between two conditions and it is calculated by. Logarithmic Fold Change.
From www.researchgate.net
Heatmap of log 2 foldchange values for ferritin (FTN), natural Logarithmic Fold Change Fold change is a metric that expresses the extent of change between two measurements. Log fold change is a statistical measure used to describe the magnitude of change in expression levels of genes between two different conditions. Log2 fold change is the ratio of expression levels between two conditions and it is calculated by taking log2 of the ratio of. Logarithmic Fold Change.
From www.researchgate.net
Log fold change of all significant and near significant metabolites Logarithmic Fold Change The log2 fold change (log2fc) term is commonly used in bioinformatics to measure the changes in gene expression between two conditions (e.g. Log2 fold change is the ratio of expression levels between two conditions and it is calculated by taking log2 of the ratio of expression levels of two conditions. In gene expression analysis, such as microarray or rna. The. Logarithmic Fold Change.
From www.researchgate.net
Foldchange in mRNA expression of SLC7A11 (A), TRIM16 (B), SRXN1 (C Logarithmic Fold Change Fold change is a metric that expresses the extent of change between two measurements. The log2 fold change (log2fc) term is commonly used in bioinformatics to measure the changes in gene expression between two conditions (e.g. We propose a fold change transform that demonstrates a combination of visualization properties exhibited by log and linear plots of fold. In gene expression. Logarithmic Fold Change.
From www.researchgate.net
Differential abundance of microbial genera (log2 fold change) (a Logarithmic Fold Change The log2 fold change (log2fc) term is commonly used in bioinformatics to measure the changes in gene expression between two conditions (e.g. Log2 fold change is the ratio of expression levels between two conditions and it is calculated by taking log2 of the ratio of expression levels of two conditions. In gene expression analysis, such as microarray or rna. The. Logarithmic Fold Change.
From www.researchgate.net
MD plot showing the logfold change and average abundance of each gene Logarithmic Fold Change Log2 fold change is the ratio of expression levels between two conditions and it is calculated by taking log2 of the ratio of expression levels of two conditions. The log2 (log with base 2) is most commonly used. The log2 fold change (log2fc) term is commonly used in bioinformatics to measure the changes in gene expression between two conditions (e.g.. Logarithmic Fold Change.
From www.researchgate.net
Heatmap indicating the average log 2 fold change in expression (yellow Logarithmic Fold Change The log2 fold change (log2fc) term is commonly used in bioinformatics to measure the changes in gene expression between two conditions (e.g. Log2 fold change values (eg 1 or 2 or 3) can be converted to fold changes by taking 2^1 or 2^2 or 2^3 = 1 or 4 or 8; The most common approach in the comparative analysis of. Logarithmic Fold Change.
From www.researchgate.net
Logarithmic fold change (Log FC) of gene expression of dynaminrelated Logarithmic Fold Change Log2 fold change values (eg 1 or 2 or 3) can be converted to fold changes by taking 2^1 or 2^2 or 2^3 = 1 or 4 or 8; In gene expression analysis, such as microarray or rna. We propose a fold change transform that demonstrates a combination of visualization properties exhibited by log and linear plots of fold. The. Logarithmic Fold Change.
From www.researchgate.net
The MAplot visualizes the logfold change in methylated read counts Logarithmic Fold Change The log2 fold change (log2fc) term is commonly used in bioinformatics to measure the changes in gene expression between two conditions (e.g. The most common approach in the comparative analysis of transcriptomics data is to test the null hypothesis that the logarithmic fold. We propose a fold change transform that demonstrates a combination of visualization properties exhibited by log and. Logarithmic Fold Change.
From www.researchgate.net
Log2 fold change scatterplot comparing genes and transcript/isoforms Logarithmic Fold Change Log fold change is a statistical measure used to describe the magnitude of change in expression levels of genes between two different conditions. Fold change is a metric that expresses the extent of change between two measurements. We propose a fold change transform that demonstrates a combination of visualization properties exhibited by log and linear plots of fold. Log2 fold. Logarithmic Fold Change.
From www.researchgate.net
Logfoldchange expression (M) versus log abundance (A) of gene Logarithmic Fold Change Log2 fold change is the ratio of expression levels between two conditions and it is calculated by taking log2 of the ratio of expression levels of two conditions. The most common approach in the comparative analysis of transcriptomics data is to test the null hypothesis that the logarithmic fold. Fold change is a metric that expresses the extent of change. Logarithmic Fold Change.
From www.researchgate.net
Logarithm of fold change (Log2FC) ratios for eight differentially Logarithmic Fold Change Log2 fold change values (eg 1 or 2 or 3) can be converted to fold changes by taking 2^1 or 2^2 or 2^3 = 1 or 4 or 8; The log2 fold change (log2fc) term is commonly used in bioinformatics to measure the changes in gene expression between two conditions (e.g. Log2 fold change is the ratio of expression levels. Logarithmic Fold Change.
From www.researchgate.net
Volcano plot. Log 2 fold change and −log 10 of the adjusted pvalue of Logarithmic Fold Change The log2 (log with base 2) is most commonly used. Log2 fold change values (eg 1 or 2 or 3) can be converted to fold changes by taking 2^1 or 2^2 or 2^3 = 1 or 4 or 8; The most common approach in the comparative analysis of transcriptomics data is to test the null hypothesis that the logarithmic fold.. Logarithmic Fold Change.
From www.researchgate.net
[A] Average logfold change of differentially expressed genes. Each Logarithmic Fold Change Log2 fold change values (eg 1 or 2 or 3) can be converted to fold changes by taking 2^1 or 2^2 or 2^3 = 1 or 4 or 8; Fold change is a metric that expresses the extent of change between two measurements. In gene expression analysis, such as microarray or rna. The log2 (log with base 2) is most. Logarithmic Fold Change.
From www.researchgate.net
Logfoldchange between the different treatments for the type I Logarithmic Fold Change The log2 fold change (log2fc) term is commonly used in bioinformatics to measure the changes in gene expression between two conditions (e.g. Fold change is a metric that expresses the extent of change between two measurements. The log2 (log with base 2) is most commonly used. In gene expression analysis, such as microarray or rna. Log2 fold change is the. Logarithmic Fold Change.
From www.researchgate.net
Volcano plot showing variation of significance and logfoldchange for Logarithmic Fold Change Log2 fold change is the ratio of expression levels between two conditions and it is calculated by taking log2 of the ratio of expression levels of two conditions. We propose a fold change transform that demonstrates a combination of visualization properties exhibited by log and linear plots of fold. The log2 (log with base 2) is most commonly used. In. Logarithmic Fold Change.
From jhudatascience.org
Chapter 6 Transformation, Foldchange, & MA Plots Statistics for Logarithmic Fold Change The most common approach in the comparative analysis of transcriptomics data is to test the null hypothesis that the logarithmic fold. Log2 fold change values (eg 1 or 2 or 3) can be converted to fold changes by taking 2^1 or 2^2 or 2^3 = 1 or 4 or 8; We propose a fold change transform that demonstrates a combination. Logarithmic Fold Change.
From www.researchgate.net
Log2 fold change in abundance of taxa associated with shifts in Logarithmic Fold Change Log2 fold change values (eg 1 or 2 or 3) can be converted to fold changes by taking 2^1 or 2^2 or 2^3 = 1 or 4 or 8; The log2 (log with base 2) is most commonly used. We propose a fold change transform that demonstrates a combination of visualization properties exhibited by log and linear plots of fold.. Logarithmic Fold Change.