Human Protein Atlas Api . The atlas for all human proteins in cells and tissues using various omics: Confocal microscopy analysis using human cell lines is performed for more detailed protein localisation. The data files represented here includes data available in the human protein atlas version 19.3. The proteins rest api provides access to key biological data from uniprot and data from large scale studies (lss) mapped to uniprot. 1463 rows data for an individual entry can be accessed in xml, rdf (trig), tsv or json format by adding the corresponding format extension. The program hosts the human. A subset of this data can also be downloaded. The human protein atlas (hpa) subcellular classification (subc) software package is a collection of python scripts by. Follow their code on github.
from www.proteinatlas.org
The human protein atlas (hpa) subcellular classification (subc) software package is a collection of python scripts by. The data files represented here includes data available in the human protein atlas version 19.3. The atlas for all human proteins in cells and tissues using various omics: Follow their code on github. The program hosts the human. Confocal microscopy analysis using human cell lines is performed for more detailed protein localisation. 1463 rows data for an individual entry can be accessed in xml, rdf (trig), tsv or json format by adding the corresponding format extension. A subset of this data can also be downloaded. The proteins rest api provides access to key biological data from uniprot and data from large scale studies (lss) mapped to uniprot.
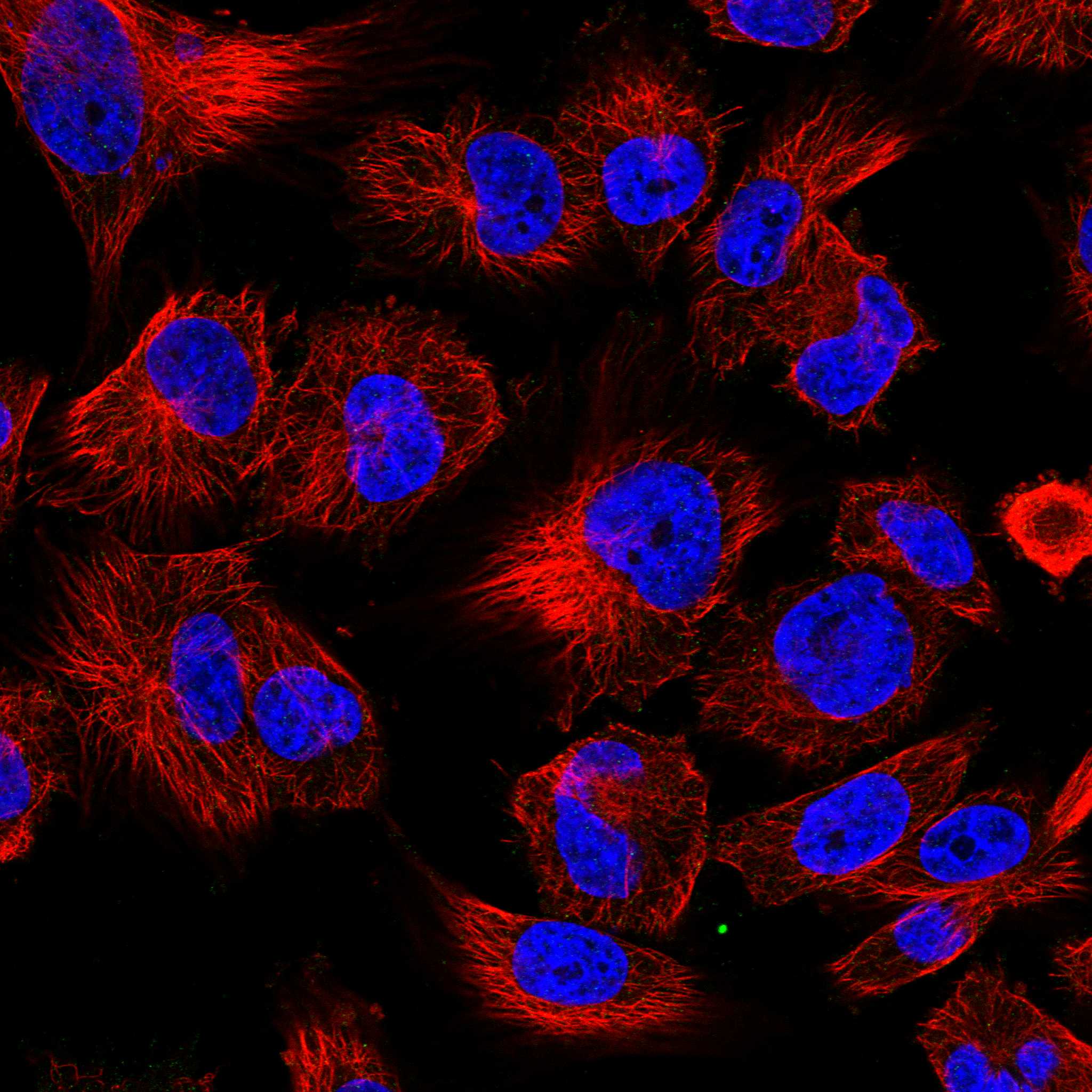
Subcellular SCUBE3 The Human Protein Atlas
Human Protein Atlas Api Confocal microscopy analysis using human cell lines is performed for more detailed protein localisation. The atlas for all human proteins in cells and tissues using various omics: The data files represented here includes data available in the human protein atlas version 19.3. The program hosts the human. The human protein atlas (hpa) subcellular classification (subc) software package is a collection of python scripts by. A subset of this data can also be downloaded. The proteins rest api provides access to key biological data from uniprot and data from large scale studies (lss) mapped to uniprot. 1463 rows data for an individual entry can be accessed in xml, rdf (trig), tsv or json format by adding the corresponding format extension. Confocal microscopy analysis using human cell lines is performed for more detailed protein localisation. Follow their code on github.
From www.embopress.org
A region‐resolved proteomic map of the human brain enabled by high Human Protein Atlas Api The human protein atlas (hpa) subcellular classification (subc) software package is a collection of python scripts by. A subset of this data can also be downloaded. The data files represented here includes data available in the human protein atlas version 19.3. 1463 rows data for an individual entry can be accessed in xml, rdf (trig), tsv or json format by. Human Protein Atlas Api.
From www.nature.com
Analysis of the Human Protein Atlas Image Classification competition Human Protein Atlas Api The proteins rest api provides access to key biological data from uniprot and data from large scale studies (lss) mapped to uniprot. The human protein atlas (hpa) subcellular classification (subc) software package is a collection of python scripts by. Confocal microscopy analysis using human cell lines is performed for more detailed protein localisation. The atlas for all human proteins in. Human Protein Atlas Api.
From www.scilifelab.se
Proteinprotein interaction networks for more than 11,000 genes in new Human Protein Atlas Api 1463 rows data for an individual entry can be accessed in xml, rdf (trig), tsv or json format by adding the corresponding format extension. The data files represented here includes data available in the human protein atlas version 19.3. The program hosts the human. A subset of this data can also be downloaded. The proteins rest api provides access to. Human Protein Atlas Api.
From play.google.com
Human Anatomy Atlas 2017 Android Apps on Google Play Human Protein Atlas Api The human protein atlas (hpa) subcellular classification (subc) software package is a collection of python scripts by. The data files represented here includes data available in the human protein atlas version 19.3. A subset of this data can also be downloaded. The program hosts the human. The atlas for all human proteins in cells and tissues using various omics: Follow. Human Protein Atlas Api.
From www.livescience.com
Human 'Atlas' Reveals Where Proteins Reside in the Body Live Science Human Protein Atlas Api Follow their code on github. Confocal microscopy analysis using human cell lines is performed for more detailed protein localisation. The data files represented here includes data available in the human protein atlas version 19.3. 1463 rows data for an individual entry can be accessed in xml, rdf (trig), tsv or json format by adding the corresponding format extension. The program. Human Protein Atlas Api.
From medevel.com
The Human Protein Atlas is What every Medical, Human Biology Students Human Protein Atlas Api 1463 rows data for an individual entry can be accessed in xml, rdf (trig), tsv or json format by adding the corresponding format extension. The human protein atlas (hpa) subcellular classification (subc) software package is a collection of python scripts by. Confocal microscopy analysis using human cell lines is performed for more detailed protein localisation. The atlas for all human. Human Protein Atlas Api.
From medevel.com
The Human Protein Atlas is What every Medical, Human Biology Students Human Protein Atlas Api Follow their code on github. The atlas for all human proteins in cells and tissues using various omics: The human protein atlas (hpa) subcellular classification (subc) software package is a collection of python scripts by. Confocal microscopy analysis using human cell lines is performed for more detailed protein localisation. A subset of this data can also be downloaded. 1463 rows. Human Protein Atlas Api.
From www.proteinatlas.org
Subcellular UNC5A The Human Protein Atlas Human Protein Atlas Api The data files represented here includes data available in the human protein atlas version 19.3. 1463 rows data for an individual entry can be accessed in xml, rdf (trig), tsv or json format by adding the corresponding format extension. The program hosts the human. A subset of this data can also be downloaded. Follow their code on github. The atlas. Human Protein Atlas Api.
From www.proteinatlas.org
Cell atlas ALOX5 The Human Protein Atlas Human Protein Atlas Api The program hosts the human. The human protein atlas (hpa) subcellular classification (subc) software package is a collection of python scripts by. 1463 rows data for an individual entry can be accessed in xml, rdf (trig), tsv or json format by adding the corresponding format extension. A subset of this data can also be downloaded. The atlas for all human. Human Protein Atlas Api.
From www.youtube.com
The Human Protein Atlas Spatial proteomics in health and disease Human Protein Atlas Api The human protein atlas (hpa) subcellular classification (subc) software package is a collection of python scripts by. The program hosts the human. Confocal microscopy analysis using human cell lines is performed for more detailed protein localisation. A subset of this data can also be downloaded. 1463 rows data for an individual entry can be accessed in xml, rdf (trig), tsv. Human Protein Atlas Api.
From www.science.org
An atlas of the proteincoding genes in the human, pig, and mouse brain Human Protein Atlas Api The human protein atlas (hpa) subcellular classification (subc) software package is a collection of python scripts by. 1463 rows data for an individual entry can be accessed in xml, rdf (trig), tsv or json format by adding the corresponding format extension. Follow their code on github. The program hosts the human. The proteins rest api provides access to key biological. Human Protein Atlas Api.
From www.researchgate.net
Human protein atlas database was used to explore six adverse expression Human Protein Atlas Api Follow their code on github. The data files represented here includes data available in the human protein atlas version 19.3. The proteins rest api provides access to key biological data from uniprot and data from large scale studies (lss) mapped to uniprot. Confocal microscopy analysis using human cell lines is performed for more detailed protein localisation. The program hosts the. Human Protein Atlas Api.
From www.proteinatlas.org
Subcellular S100A14 The Human Protein Atlas Human Protein Atlas Api The atlas for all human proteins in cells and tissues using various omics: A subset of this data can also be downloaded. The program hosts the human. Confocal microscopy analysis using human cell lines is performed for more detailed protein localisation. Follow their code on github. The human protein atlas (hpa) subcellular classification (subc) software package is a collection of. Human Protein Atlas Api.
From www.researchgate.net
The Human Protein Atlas. This portal contains Human Protein Atlas Api The data files represented here includes data available in the human protein atlas version 19.3. The atlas for all human proteins in cells and tissues using various omics: The human protein atlas (hpa) subcellular classification (subc) software package is a collection of python scripts by. Follow their code on github. 1463 rows data for an individual entry can be accessed. Human Protein Atlas Api.
From www.researchgate.net
Analysis of the Human Protein Atlas Image Classification competition Human Protein Atlas Api 1463 rows data for an individual entry can be accessed in xml, rdf (trig), tsv or json format by adding the corresponding format extension. The atlas for all human proteins in cells and tissues using various omics: The program hosts the human. Confocal microscopy analysis using human cell lines is performed for more detailed protein localisation. The human protein atlas. Human Protein Atlas Api.
From www.proteinatlas.org
Subcellular AIM2 The Human Protein Atlas Human Protein Atlas Api The human protein atlas (hpa) subcellular classification (subc) software package is a collection of python scripts by. The program hosts the human. The atlas for all human proteins in cells and tissues using various omics: A subset of this data can also be downloaded. Confocal microscopy analysis using human cell lines is performed for more detailed protein localisation. Follow their. Human Protein Atlas Api.
From www.nature.com
Analysis of the Human Protein Atlas Image Classification competition Human Protein Atlas Api The atlas for all human proteins in cells and tissues using various omics: Confocal microscopy analysis using human cell lines is performed for more detailed protein localisation. Follow their code on github. 1463 rows data for an individual entry can be accessed in xml, rdf (trig), tsv or json format by adding the corresponding format extension. The data files represented. Human Protein Atlas Api.
From bmcmedicine.biomedcentral.com
A tool to facilitate clinical biomarker studies a tissue dictionary Human Protein Atlas Api 1463 rows data for an individual entry can be accessed in xml, rdf (trig), tsv or json format by adding the corresponding format extension. Confocal microscopy analysis using human cell lines is performed for more detailed protein localisation. Follow their code on github. The data files represented here includes data available in the human protein atlas version 19.3. The atlas. Human Protein Atlas Api.
From www.youtube.com
Using Human Protein Atlas to Find Tissue Specific Genes in R using Human Protein Atlas Api The data files represented here includes data available in the human protein atlas version 19.3. The proteins rest api provides access to key biological data from uniprot and data from large scale studies (lss) mapped to uniprot. The atlas for all human proteins in cells and tissues using various omics: Follow their code on github. 1463 rows data for an. Human Protein Atlas Api.
From www.scilifelab.se
The Human protein Atlas celebrates 20 years SciLifeLab Human Protein Atlas Api The proteins rest api provides access to key biological data from uniprot and data from large scale studies (lss) mapped to uniprot. The human protein atlas (hpa) subcellular classification (subc) software package is a collection of python scripts by. The data files represented here includes data available in the human protein atlas version 19.3. The atlas for all human proteins. Human Protein Atlas Api.
From www.atlasantibodies.com
New Triple A Polyclonals Human Protein Atlas release 2023 Human Protein Atlas Api The proteins rest api provides access to key biological data from uniprot and data from large scale studies (lss) mapped to uniprot. A subset of this data can also be downloaded. The human protein atlas (hpa) subcellular classification (subc) software package is a collection of python scripts by. 1463 rows data for an individual entry can be accessed in xml,. Human Protein Atlas Api.
From v15.proteinatlas.org
ESR1 Antibodies The Human Protein Atlas Human Protein Atlas Api The data files represented here includes data available in the human protein atlas version 19.3. A subset of this data can also be downloaded. The human protein atlas (hpa) subcellular classification (subc) software package is a collection of python scripts by. The program hosts the human. Follow their code on github. 1463 rows data for an individual entry can be. Human Protein Atlas Api.
From www.proteinatlas.org
Subcellular SCUBE3 The Human Protein Atlas Human Protein Atlas Api Confocal microscopy analysis using human cell lines is performed for more detailed protein localisation. The data files represented here includes data available in the human protein atlas version 19.3. A subset of this data can also be downloaded. The human protein atlas (hpa) subcellular classification (subc) software package is a collection of python scripts by. The proteins rest api provides. Human Protein Atlas Api.
From libraryguides.ccbcmd.edu
Digital Histology Histotechnology Research Guides at Community Human Protein Atlas Api A subset of this data can also be downloaded. Confocal microscopy analysis using human cell lines is performed for more detailed protein localisation. The human protein atlas (hpa) subcellular classification (subc) software package is a collection of python scripts by. The atlas for all human proteins in cells and tissues using various omics: 1463 rows data for an individual entry. Human Protein Atlas Api.
From www.science.org
Evolution and antiviral activity of a human protein of retroviral Human Protein Atlas Api The human protein atlas (hpa) subcellular classification (subc) software package is a collection of python scripts by. The proteins rest api provides access to key biological data from uniprot and data from large scale studies (lss) mapped to uniprot. The data files represented here includes data available in the human protein atlas version 19.3. 1463 rows data for an individual. Human Protein Atlas Api.
From www.ge.com
Human Protein Atlas GE News Human Protein Atlas Api The proteins rest api provides access to key biological data from uniprot and data from large scale studies (lss) mapped to uniprot. The human protein atlas (hpa) subcellular classification (subc) software package is a collection of python scripts by. Follow their code on github. Confocal microscopy analysis using human cell lines is performed for more detailed protein localisation. A subset. Human Protein Atlas Api.
From www.scilifelab.se
The Human Protein Atlas reaches a major milestone SciLifeLab Human Protein Atlas Api The human protein atlas (hpa) subcellular classification (subc) software package is a collection of python scripts by. The atlas for all human proteins in cells and tissues using various omics: Follow their code on github. A subset of this data can also be downloaded. 1463 rows data for an individual entry can be accessed in xml, rdf (trig), tsv or. Human Protein Atlas Api.
From www.researchgate.net
Human Protein Atlas immunohistochemistry using antiEgr3 antibodies Human Protein Atlas Api The atlas for all human proteins in cells and tissues using various omics: The proteins rest api provides access to key biological data from uniprot and data from large scale studies (lss) mapped to uniprot. The data files represented here includes data available in the human protein atlas version 19.3. The human protein atlas (hpa) subcellular classification (subc) software package. Human Protein Atlas Api.
From peerj.com
Integrated analysis of the roles and prognostic value of RNA binding Human Protein Atlas Api A subset of this data can also be downloaded. The program hosts the human. 1463 rows data for an individual entry can be accessed in xml, rdf (trig), tsv or json format by adding the corresponding format extension. Confocal microscopy analysis using human cell lines is performed for more detailed protein localisation. The human protein atlas (hpa) subcellular classification (subc). Human Protein Atlas Api.
From onlinelibrary.wiley.com
The human protein atlas A spatial map of the human proteome Thul Human Protein Atlas Api Follow their code on github. 1463 rows data for an individual entry can be accessed in xml, rdf (trig), tsv or json format by adding the corresponding format extension. The human protein atlas (hpa) subcellular classification (subc) software package is a collection of python scripts by. The proteins rest api provides access to key biological data from uniprot and data. Human Protein Atlas Api.
From www.proteinatlas.org
Subcellular MTARC1 The Human Protein Atlas Human Protein Atlas Api Follow their code on github. A subset of this data can also be downloaded. The data files represented here includes data available in the human protein atlas version 19.3. The atlas for all human proteins in cells and tissues using various omics: Confocal microscopy analysis using human cell lines is performed for more detailed protein localisation. The proteins rest api. Human Protein Atlas Api.
From www.researchgate.net
The Protein Atlas concept. Download Scientific Diagram Human Protein Atlas Api The data files represented here includes data available in the human protein atlas version 19.3. The atlas for all human proteins in cells and tissues using various omics: The program hosts the human. The human protein atlas (hpa) subcellular classification (subc) software package is a collection of python scripts by. A subset of this data can also be downloaded. 1463. Human Protein Atlas Api.
From www.proteinatlas.org
Subcellular ITPRID2 The Human Protein Atlas Human Protein Atlas Api A subset of this data can also be downloaded. The data files represented here includes data available in the human protein atlas version 19.3. The atlas for all human proteins in cells and tissues using various omics: Confocal microscopy analysis using human cell lines is performed for more detailed protein localisation. 1463 rows data for an individual entry can be. Human Protein Atlas Api.
From www.proteinatlas.org
Subcellular COL1A2 The Human Protein Atlas Human Protein Atlas Api 1463 rows data for an individual entry can be accessed in xml, rdf (trig), tsv or json format by adding the corresponding format extension. The program hosts the human. Confocal microscopy analysis using human cell lines is performed for more detailed protein localisation. A subset of this data can also be downloaded. The proteins rest api provides access to key. Human Protein Atlas Api.
From www.proteinatlas.org
Subcellular LSM2 The Human Protein Atlas Human Protein Atlas Api The proteins rest api provides access to key biological data from uniprot and data from large scale studies (lss) mapped to uniprot. The data files represented here includes data available in the human protein atlas version 19.3. Follow their code on github. Confocal microscopy analysis using human cell lines is performed for more detailed protein localisation. A subset of this. Human Protein Atlas Api.