Differential Gene Expression Analysis Using R . This course is an introduction to differential expression analysis from rnaseq data. Import gene count and meta data. For each gene in each sample, normalise by dividing by the geometric mean. This includes reading the data into r, quality control and preprocessing, and. Set up the deseqdataset, run the. Limma is an r package that was originally developed for differential expression (de) analysis of gene expression microarray data. First, import the countdata and metadata directly from the web. It will take you from the raw fastq files all the way to the. For each gene calculate the geometric mean across all samples.
from www.spandidos-publications.com
Limma is an r package that was originally developed for differential expression (de) analysis of gene expression microarray data. Set up the deseqdataset, run the. For each gene calculate the geometric mean across all samples. It will take you from the raw fastq files all the way to the. This includes reading the data into r, quality control and preprocessing, and. Import gene count and meta data. This course is an introduction to differential expression analysis from rnaseq data. First, import the countdata and metadata directly from the web. For each gene in each sample, normalise by dividing by the geometric mean.
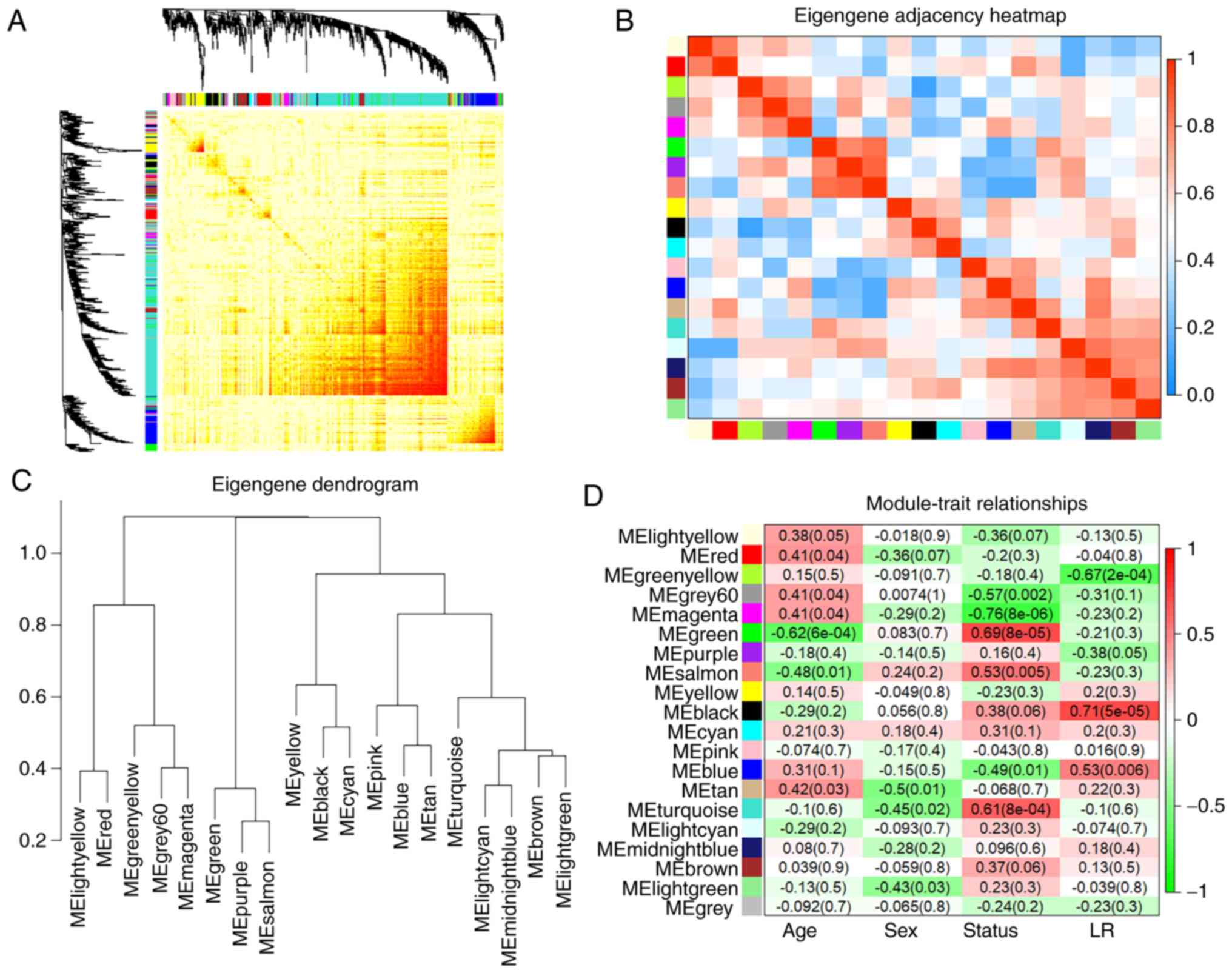
Weighted gene co‑expression network analysis to identify key modules
Differential Gene Expression Analysis Using R Import gene count and meta data. This includes reading the data into r, quality control and preprocessing, and. For each gene in each sample, normalise by dividing by the geometric mean. First, import the countdata and metadata directly from the web. Limma is an r package that was originally developed for differential expression (de) analysis of gene expression microarray data. For each gene calculate the geometric mean across all samples. Import gene count and meta data. This course is an introduction to differential expression analysis from rnaseq data. Set up the deseqdataset, run the. It will take you from the raw fastq files all the way to the.
From ar.inspiredpencil.com
Expressed Differential Gene Expression Analysis Using R First, import the countdata and metadata directly from the web. Import gene count and meta data. It will take you from the raw fastq files all the way to the. For each gene calculate the geometric mean across all samples. Limma is an r package that was originally developed for differential expression (de) analysis of gene expression microarray data. This. Differential Gene Expression Analysis Using R.
From www.vrogue.co
Heatmap Differential Gene Expression Analysis Biocode vrogue.co Differential Gene Expression Analysis Using R For each gene in each sample, normalise by dividing by the geometric mean. For each gene calculate the geometric mean across all samples. Limma is an r package that was originally developed for differential expression (de) analysis of gene expression microarray data. It will take you from the raw fastq files all the way to the. First, import the countdata. Differential Gene Expression Analysis Using R.
From carlsbattles.blob.core.windows.net
Gene Expression Vs Transcription at carlsbattles blog Differential Gene Expression Analysis Using R This course is an introduction to differential expression analysis from rnaseq data. Import gene count and meta data. Set up the deseqdataset, run the. For each gene calculate the geometric mean across all samples. For each gene in each sample, normalise by dividing by the geometric mean. First, import the countdata and metadata directly from the web. Limma is an. Differential Gene Expression Analysis Using R.
From www.researchgate.net
Scheme of the methods employed for differential gene expression Differential Gene Expression Analysis Using R Set up the deseqdataset, run the. It will take you from the raw fastq files all the way to the. For each gene in each sample, normalise by dividing by the geometric mean. This course is an introduction to differential expression analysis from rnaseq data. This includes reading the data into r, quality control and preprocessing, and. For each gene. Differential Gene Expression Analysis Using R.
From www.sexizpix.com
Heatmap Of Differential Gene Expression Correlation Across All Sexiz Pix Differential Gene Expression Analysis Using R Limma is an r package that was originally developed for differential expression (de) analysis of gene expression microarray data. It will take you from the raw fastq files all the way to the. First, import the countdata and metadata directly from the web. This course is an introduction to differential expression analysis from rnaseq data. Set up the deseqdataset, run. Differential Gene Expression Analysis Using R.
From hgserver1.amc.nl
R2 Genomics Analysis and Visualization Platform Differential Gene Expression Analysis Using R Set up the deseqdataset, run the. This includes reading the data into r, quality control and preprocessing, and. First, import the countdata and metadata directly from the web. For each gene in each sample, normalise by dividing by the geometric mean. This course is an introduction to differential expression analysis from rnaseq data. Import gene count and meta data. It. Differential Gene Expression Analysis Using R.
From www.vrogue.co
Heatmap Differential Gene Expression Analysis Biocode vrogue.co Differential Gene Expression Analysis Using R Set up the deseqdataset, run the. Import gene count and meta data. For each gene calculate the geometric mean across all samples. This course is an introduction to differential expression analysis from rnaseq data. First, import the countdata and metadata directly from the web. Limma is an r package that was originally developed for differential expression (de) analysis of gene. Differential Gene Expression Analysis Using R.
From www.spandidos-publications.com
Weighted gene co‑expression network analysis to identify key modules Differential Gene Expression Analysis Using R First, import the countdata and metadata directly from the web. For each gene in each sample, normalise by dividing by the geometric mean. For each gene calculate the geometric mean across all samples. Limma is an r package that was originally developed for differential expression (de) analysis of gene expression microarray data. It will take you from the raw fastq. Differential Gene Expression Analysis Using R.
From www.researchgate.net
Quantitative proteomics and KEGG pathway enrichment analysis. a Differential Gene Expression Analysis Using R Limma is an r package that was originally developed for differential expression (de) analysis of gene expression microarray data. Import gene count and meta data. For each gene calculate the geometric mean across all samples. First, import the countdata and metadata directly from the web. Set up the deseqdataset, run the. For each gene in each sample, normalise by dividing. Differential Gene Expression Analysis Using R.
From www.researchgate.net
Differential gene expression analysis. (a) Genes differentially up and Differential Gene Expression Analysis Using R It will take you from the raw fastq files all the way to the. For each gene in each sample, normalise by dividing by the geometric mean. For each gene calculate the geometric mean across all samples. This includes reading the data into r, quality control and preprocessing, and. This course is an introduction to differential expression analysis from rnaseq. Differential Gene Expression Analysis Using R.
From www.slideserve.com
PPT Gene Regulation results in differential Gene Expression, leading Differential Gene Expression Analysis Using R Set up the deseqdataset, run the. It will take you from the raw fastq files all the way to the. Import gene count and meta data. This course is an introduction to differential expression analysis from rnaseq data. For each gene calculate the geometric mean across all samples. First, import the countdata and metadata directly from the web. Limma is. Differential Gene Expression Analysis Using R.
From www.frontiersin.org
Frontiers Identification of Key Genes With Differential Correlations Differential Gene Expression Analysis Using R For each gene in each sample, normalise by dividing by the geometric mean. Limma is an r package that was originally developed for differential expression (de) analysis of gene expression microarray data. This course is an introduction to differential expression analysis from rnaseq data. It will take you from the raw fastq files all the way to the. Import gene. Differential Gene Expression Analysis Using R.
From www.researchgate.net
RNAseq and DEG analysis. (A) Comparison of FLNB expression levels Differential Gene Expression Analysis Using R For each gene in each sample, normalise by dividing by the geometric mean. Limma is an r package that was originally developed for differential expression (de) analysis of gene expression microarray data. Import gene count and meta data. Set up the deseqdataset, run the. This includes reading the data into r, quality control and preprocessing, and. This course is an. Differential Gene Expression Analysis Using R.
From peerj.com
Differential gene expression analysis by RNAseq reveals the importance Differential Gene Expression Analysis Using R It will take you from the raw fastq files all the way to the. Set up the deseqdataset, run the. This course is an introduction to differential expression analysis from rnaseq data. First, import the countdata and metadata directly from the web. This includes reading the data into r, quality control and preprocessing, and. For each gene calculate the geometric. Differential Gene Expression Analysis Using R.
From www.researchgate.net
Gene expression correlation between RTqPCR and RNAseq data. The Differential Gene Expression Analysis Using R Limma is an r package that was originally developed for differential expression (de) analysis of gene expression microarray data. For each gene in each sample, normalise by dividing by the geometric mean. Import gene count and meta data. First, import the countdata and metadata directly from the web. It will take you from the raw fastq files all the way. Differential Gene Expression Analysis Using R.
From scienceparkstudygroup.github.io
06 Differential expression analysis Introduction to RNAseq Differential Gene Expression Analysis Using R For each gene calculate the geometric mean across all samples. First, import the countdata and metadata directly from the web. Set up the deseqdataset, run the. This includes reading the data into r, quality control and preprocessing, and. Limma is an r package that was originally developed for differential expression (de) analysis of gene expression microarray data. This course is. Differential Gene Expression Analysis Using R.
From www.researchgate.net
Differential expression analysis shows two sets of mutants with Differential Gene Expression Analysis Using R It will take you from the raw fastq files all the way to the. For each gene in each sample, normalise by dividing by the geometric mean. Import gene count and meta data. First, import the countdata and metadata directly from the web. This course is an introduction to differential expression analysis from rnaseq data. Set up the deseqdataset, run. Differential Gene Expression Analysis Using R.
From www.researchgate.net
Clustering of geneexpression data. (A) Unsupervised hierarchical Differential Gene Expression Analysis Using R Limma is an r package that was originally developed for differential expression (de) analysis of gene expression microarray data. First, import the countdata and metadata directly from the web. For each gene in each sample, normalise by dividing by the geometric mean. Import gene count and meta data. Set up the deseqdataset, run the. This includes reading the data into. Differential Gene Expression Analysis Using R.
From www.researchgate.net
Differential gene expression analysis and GSEA of the scRNAseq Differential Gene Expression Analysis Using R This includes reading the data into r, quality control and preprocessing, and. First, import the countdata and metadata directly from the web. It will take you from the raw fastq files all the way to the. This course is an introduction to differential expression analysis from rnaseq data. Import gene count and meta data. For each gene in each sample,. Differential Gene Expression Analysis Using R.
From bitesizebio.com
Show Disparity in Gene Expression with a Heat Map Differential Gene Expression Analysis Using R Set up the deseqdataset, run the. This course is an introduction to differential expression analysis from rnaseq data. Limma is an r package that was originally developed for differential expression (de) analysis of gene expression microarray data. It will take you from the raw fastq files all the way to the. For each gene in each sample, normalise by dividing. Differential Gene Expression Analysis Using R.
From hbctraining.github.io
Count normalization with DESeq2 Introduction to DGE ARCHIVED Differential Gene Expression Analysis Using R First, import the countdata and metadata directly from the web. Limma is an r package that was originally developed for differential expression (de) analysis of gene expression microarray data. Import gene count and meta data. For each gene in each sample, normalise by dividing by the geometric mean. For each gene calculate the geometric mean across all samples. This course. Differential Gene Expression Analysis Using R.