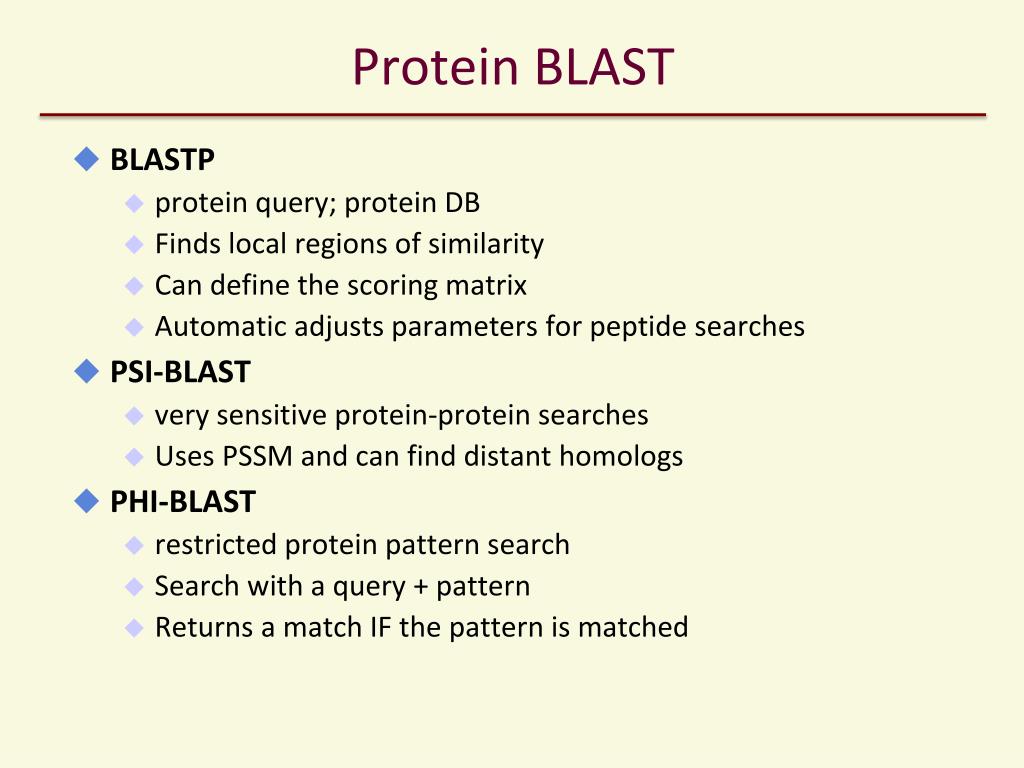
Protein Blast Two Sequences . blastp (protein blast): this episode of the blast tutorial series demonstrates how to. Then use the blast button. its speed and sensitivity allow scientists to compare nucleotide and protein sequences to both single sequences and large. enter one or more queries in the top text box and one or more subject sequences in the lower text box. the basic local alignment search tool (blast) finds regions of local similarity between sequences. a global algorithm returns one alignment clearly showing the difference, a local algorithm returns two alignments, and it is. Compares one or more protein query sequences to a subject. the basic local alignment search tool (blast) finds regions of local similarity between protein or. comparing two or more protein sequences using blastp. The program compares nucleotide or protein.
from www.slideserve.com
this episode of the blast tutorial series demonstrates how to. Then use the blast button. enter one or more queries in the top text box and one or more subject sequences in the lower text box. Compares one or more protein query sequences to a subject. blastp (protein blast): comparing two or more protein sequences using blastp. The program compares nucleotide or protein. a global algorithm returns one alignment clearly showing the difference, a local algorithm returns two alignments, and it is. the basic local alignment search tool (blast) finds regions of local similarity between protein or. its speed and sensitivity allow scientists to compare nucleotide and protein sequences to both single sequences and large.
PPT Sequence Similarity & Alignment with BLAST PowerPoint
Protein Blast Two Sequences enter one or more queries in the top text box and one or more subject sequences in the lower text box. its speed and sensitivity allow scientists to compare nucleotide and protein sequences to both single sequences and large. a global algorithm returns one alignment clearly showing the difference, a local algorithm returns two alignments, and it is. Compares one or more protein query sequences to a subject. Then use the blast button. comparing two or more protein sequences using blastp. the basic local alignment search tool (blast) finds regions of local similarity between sequences. blastp (protein blast): enter one or more queries in the top text box and one or more subject sequences in the lower text box. The program compares nucleotide or protein. this episode of the blast tutorial series demonstrates how to. the basic local alignment search tool (blast) finds regions of local similarity between protein or.
From www.news-medical.net
Computational study reveals SARSCoV2 spike protein structural stability Protein Blast Two Sequences enter one or more queries in the top text box and one or more subject sequences in the lower text box. the basic local alignment search tool (blast) finds regions of local similarity between sequences. this episode of the blast tutorial series demonstrates how to. blastp (protein blast): a global algorithm returns one alignment clearly. Protein Blast Two Sequences.
From medium.com
Using the NCBI BLAST Interface For Identifying Homologous Sequences In Protein Blast Two Sequences a global algorithm returns one alignment clearly showing the difference, a local algorithm returns two alignments, and it is. the basic local alignment search tool (blast) finds regions of local similarity between protein or. The program compares nucleotide or protein. enter one or more queries in the top text box and one or more subject sequences in. Protein Blast Two Sequences.
From abacus.bates.edu
BLASTP Results Part 1 Protein Blast Two Sequences Compares one or more protein query sequences to a subject. The program compares nucleotide or protein. the basic local alignment search tool (blast) finds regions of local similarity between protein or. comparing two or more protein sequences using blastp. the basic local alignment search tool (blast) finds regions of local similarity between sequences. a global algorithm. Protein Blast Two Sequences.
From www.slideserve.com
PPT Molecular Evidence PowerPoint Presentation, free download ID595756 Protein Blast Two Sequences a global algorithm returns one alignment clearly showing the difference, a local algorithm returns two alignments, and it is. blastp (protein blast): its speed and sensitivity allow scientists to compare nucleotide and protein sequences to both single sequences and large. enter one or more queries in the top text box and one or more subject sequences. Protein Blast Two Sequences.
From giovzxsoi.blob.core.windows.net
Examples Of Proteins In Biology at Carlos Collins blog Protein Blast Two Sequences comparing two or more protein sequences using blastp. enter one or more queries in the top text box and one or more subject sequences in the lower text box. this episode of the blast tutorial series demonstrates how to. a global algorithm returns one alignment clearly showing the difference, a local algorithm returns two alignments, and. Protein Blast Two Sequences.
From support.nlm.nih.gov
How do I interpret Nucleotide BLAST (blastn) pairwise alignments with Protein Blast Two Sequences this episode of the blast tutorial series demonstrates how to. its speed and sensitivity allow scientists to compare nucleotide and protein sequences to both single sequences and large. enter one or more queries in the top text box and one or more subject sequences in the lower text box. The program compares nucleotide or protein. the. Protein Blast Two Sequences.
From www.youtube.com
Which BLAST Algorithm to use for ProteinProtein BLAST YouTube Protein Blast Two Sequences comparing two or more protein sequences using blastp. the basic local alignment search tool (blast) finds regions of local similarity between protein or. blastp (protein blast): enter one or more queries in the top text box and one or more subject sequences in the lower text box. The program compares nucleotide or protein. its speed. Protein Blast Two Sequences.
From www.youtube.com
Translated Nucleotide to Protein BLAST BLASTX YouTube Protein Blast Two Sequences the basic local alignment search tool (blast) finds regions of local similarity between sequences. this episode of the blast tutorial series demonstrates how to. a global algorithm returns one alignment clearly showing the difference, a local algorithm returns two alignments, and it is. comparing two or more protein sequences using blastp. enter one or more. Protein Blast Two Sequences.
From www.slideserve.com
PPT Pairwise and Multiple Sequence Alignment Lesson 2 PowerPoint Protein Blast Two Sequences its speed and sensitivity allow scientists to compare nucleotide and protein sequences to both single sequences and large. comparing two or more protein sequences using blastp. the basic local alignment search tool (blast) finds regions of local similarity between protein or. The program compares nucleotide or protein. Then use the blast button. the basic local alignment. Protein Blast Two Sequences.
From www.researchgate.net
Multiple protein sequence alignment of HTT exon 29. Multiple protein Protein Blast Two Sequences the basic local alignment search tool (blast) finds regions of local similarity between sequences. blastp (protein blast): the basic local alignment search tool (blast) finds regions of local similarity between protein or. enter one or more queries in the top text box and one or more subject sequences in the lower text box. comparing two. Protein Blast Two Sequences.
From pubs.acs.org
Mathematical Approach to Protein Sequence Comparison Based on Protein Blast Two Sequences enter one or more queries in the top text box and one or more subject sequences in the lower text box. comparing two or more protein sequences using blastp. the basic local alignment search tool (blast) finds regions of local similarity between protein or. The program compares nucleotide or protein. Compares one or more protein query sequences. Protein Blast Two Sequences.
From support.nlm.nih.gov
How do I use Nucleotide BLAST (blastn) and CDS feature display to Protein Blast Two Sequences Compares one or more protein query sequences to a subject. this episode of the blast tutorial series demonstrates how to. a global algorithm returns one alignment clearly showing the difference, a local algorithm returns two alignments, and it is. enter one or more queries in the top text box and one or more subject sequences in the. Protein Blast Two Sequences.
From www.researchgate.net
BLAST analysis of protein sequence alignment between collagen Protein Blast Two Sequences enter one or more queries in the top text box and one or more subject sequences in the lower text box. The program compares nucleotide or protein. comparing two or more protein sequences using blastp. Compares one or more protein query sequences to a subject. the basic local alignment search tool (blast) finds regions of local similarity. Protein Blast Two Sequences.
From www.biologyexams4u.com
Importance of Sequence Alignment in Bioinformatics Biology Exams 4 U Protein Blast Two Sequences comparing two or more protein sequences using blastp. blastp (protein blast): this episode of the blast tutorial series demonstrates how to. Then use the blast button. a global algorithm returns one alignment clearly showing the difference, a local algorithm returns two alignments, and it is. enter one or more queries in the top text box. Protein Blast Two Sequences.
From usuhs.libguides.com
BLASTP Protein Sequence Search Introduction to NCBI Bioinformatics Protein Blast Two Sequences the basic local alignment search tool (blast) finds regions of local similarity between sequences. a global algorithm returns one alignment clearly showing the difference, a local algorithm returns two alignments, and it is. Then use the blast button. blastp (protein blast): this episode of the blast tutorial series demonstrates how to. Compares one or more protein. Protein Blast Two Sequences.
From www.creative-biostructure.com
Protein Sequence Analysis for Coronavirus Research Creative Protein Blast Two Sequences The program compares nucleotide or protein. a global algorithm returns one alignment clearly showing the difference, a local algorithm returns two alignments, and it is. enter one or more queries in the top text box and one or more subject sequences in the lower text box. the basic local alignment search tool (blast) finds regions of local. Protein Blast Two Sequences.
From sequenceserver.com
Identifying gene sequences in a new genome assembly using BLAST Protein Blast Two Sequences blastp (protein blast): comparing two or more protein sequences using blastp. The program compares nucleotide or protein. its speed and sensitivity allow scientists to compare nucleotide and protein sequences to both single sequences and large. Then use the blast button. this episode of the blast tutorial series demonstrates how to. the basic local alignment search. Protein Blast Two Sequences.
From www.researchgate.net
BLASTP protein comparison of the annotated sequence BC000036325 Protein Blast Two Sequences Compares one or more protein query sequences to a subject. comparing two or more protein sequences using blastp. this episode of the blast tutorial series demonstrates how to. The program compares nucleotide or protein. its speed and sensitivity allow scientists to compare nucleotide and protein sequences to both single sequences and large. blastp (protein blast): Then. Protein Blast Two Sequences.
From www.slideserve.com
PPT Protein Sequence PowerPoint Presentation, free download ID4271810 Protein Blast Two Sequences this episode of the blast tutorial series demonstrates how to. blastp (protein blast): the basic local alignment search tool (blast) finds regions of local similarity between sequences. comparing two or more protein sequences using blastp. Compares one or more protein query sequences to a subject. the basic local alignment search tool (blast) finds regions of. Protein Blast Two Sequences.
From www.ucl.ac.uk
BLAST Protein Blast Two Sequences a global algorithm returns one alignment clearly showing the difference, a local algorithm returns two alignments, and it is. Compares one or more protein query sequences to a subject. its speed and sensitivity allow scientists to compare nucleotide and protein sequences to both single sequences and large. the basic local alignment search tool (blast) finds regions of. Protein Blast Two Sequences.
From exyenvloh.blob.core.windows.net
Dna/Protein Sequences at Rodney Melville blog Protein Blast Two Sequences The program compares nucleotide or protein. comparing two or more protein sequences using blastp. its speed and sensitivity allow scientists to compare nucleotide and protein sequences to both single sequences and large. the basic local alignment search tool (blast) finds regions of local similarity between protein or. the basic local alignment search tool (blast) finds regions. Protein Blast Two Sequences.
From support.nlm.nih.gov
How do I use Nucleotide BLAST (blastn) to determine the coding Protein Blast Two Sequences Then use the blast button. Compares one or more protein query sequences to a subject. the basic local alignment search tool (blast) finds regions of local similarity between sequences. comparing two or more protein sequences using blastp. a global algorithm returns one alignment clearly showing the difference, a local algorithm returns two alignments, and it is. . Protein Blast Two Sequences.
From www.semanticscholar.org
Figure 1 from BLAST 2 Sequences, a new tool for comparing protein and Protein Blast Two Sequences the basic local alignment search tool (blast) finds regions of local similarity between protein or. a global algorithm returns one alignment clearly showing the difference, a local algorithm returns two alignments, and it is. comparing two or more protein sequences using blastp. blastp (protein blast): its speed and sensitivity allow scientists to compare nucleotide and. Protein Blast Two Sequences.
From aaronconnolly.z19.web.core.windows.net
Amino Acid Protein Chart Protein Blast Two Sequences enter one or more queries in the top text box and one or more subject sequences in the lower text box. The program compares nucleotide or protein. Then use the blast button. this episode of the blast tutorial series demonstrates how to. blastp (protein blast): Compares one or more protein query sequences to a subject. the. Protein Blast Two Sequences.
From www.slideserve.com
PPT Sequence Alignment PowerPoint Presentation, free download ID Protein Blast Two Sequences this episode of the blast tutorial series demonstrates how to. blastp (protein blast): its speed and sensitivity allow scientists to compare nucleotide and protein sequences to both single sequences and large. a global algorithm returns one alignment clearly showing the difference, a local algorithm returns two alignments, and it is. Then use the blast button. The. Protein Blast Two Sequences.
From www.youtube.com
How to Use BLAST for Finding and Aligning DNA or Protein Sequences Protein Blast Two Sequences Compares one or more protein query sequences to a subject. this episode of the blast tutorial series demonstrates how to. a global algorithm returns one alignment clearly showing the difference, a local algorithm returns two alignments, and it is. blastp (protein blast): its speed and sensitivity allow scientists to compare nucleotide and protein sequences to both. Protein Blast Two Sequences.
From www.researchgate.net
Protein sequence and structure alignment of JAK family PTK domain. (A Protein Blast Two Sequences Compares one or more protein query sequences to a subject. the basic local alignment search tool (blast) finds regions of local similarity between protein or. enter one or more queries in the top text box and one or more subject sequences in the lower text box. blastp (protein blast): the basic local alignment search tool (blast). Protein Blast Two Sequences.
From www.cell.com
A “Whirly” Transcription Factor Is Required for Salicylic Acid Protein Blast Two Sequences the basic local alignment search tool (blast) finds regions of local similarity between protein or. enter one or more queries in the top text box and one or more subject sequences in the lower text box. blastp (protein blast): Compares one or more protein query sequences to a subject. comparing two or more protein sequences using. Protein Blast Two Sequences.
From guides.nnlm.gov
Guide on the Side NCBI BLAST (Part A) Identifying Sequences Single Protein Blast Two Sequences the basic local alignment search tool (blast) finds regions of local similarity between protein or. enter one or more queries in the top text box and one or more subject sequences in the lower text box. this episode of the blast tutorial series demonstrates how to. a global algorithm returns one alignment clearly showing the difference,. Protein Blast Two Sequences.
From answerhappy.com
Blast Search Below are two protein sequences in FASTA format. Perform a Protein Blast Two Sequences blastp (protein blast): Then use the blast button. this episode of the blast tutorial series demonstrates how to. comparing two or more protein sequences using blastp. the basic local alignment search tool (blast) finds regions of local similarity between sequences. enter one or more queries in the top text box and one or more subject. Protein Blast Two Sequences.
From www.researchgate.net
The main framework of HBLAST and the workflow on the protein sequence Protein Blast Two Sequences this episode of the blast tutorial series demonstrates how to. the basic local alignment search tool (blast) finds regions of local similarity between sequences. enter one or more queries in the top text box and one or more subject sequences in the lower text box. The program compares nucleotide or protein. blastp (protein blast): its. Protein Blast Two Sequences.
From www.mun.ca
Positive and Negative Regulation Protein Blast Two Sequences Then use the blast button. the basic local alignment search tool (blast) finds regions of local similarity between protein or. the basic local alignment search tool (blast) finds regions of local similarity between sequences. its speed and sensitivity allow scientists to compare nucleotide and protein sequences to both single sequences and large. enter one or more. Protein Blast Two Sequences.
From support.nlm.nih.gov
How do I use Nucleotide BLAST (blastn) with CDS feature display to Protein Blast Two Sequences the basic local alignment search tool (blast) finds regions of local similarity between sequences. a global algorithm returns one alignment clearly showing the difference, a local algorithm returns two alignments, and it is. Then use the blast button. this episode of the blast tutorial series demonstrates how to. Compares one or more protein query sequences to a. Protein Blast Two Sequences.
From www.slideserve.com
PPT Sequence Similarity & Alignment with BLAST PowerPoint Protein Blast Two Sequences its speed and sensitivity allow scientists to compare nucleotide and protein sequences to both single sequences and large. the basic local alignment search tool (blast) finds regions of local similarity between protein or. comparing two or more protein sequences using blastp. blastp (protein blast): a global algorithm returns one alignment clearly showing the difference, a. Protein Blast Two Sequences.
From www.slideserve.com
PPT NCBI PowerPoint Presentation ID315849 Protein Blast Two Sequences this episode of the blast tutorial series demonstrates how to. Then use the blast button. enter one or more queries in the top text box and one or more subject sequences in the lower text box. a global algorithm returns one alignment clearly showing the difference, a local algorithm returns two alignments, and it is. the. Protein Blast Two Sequences.