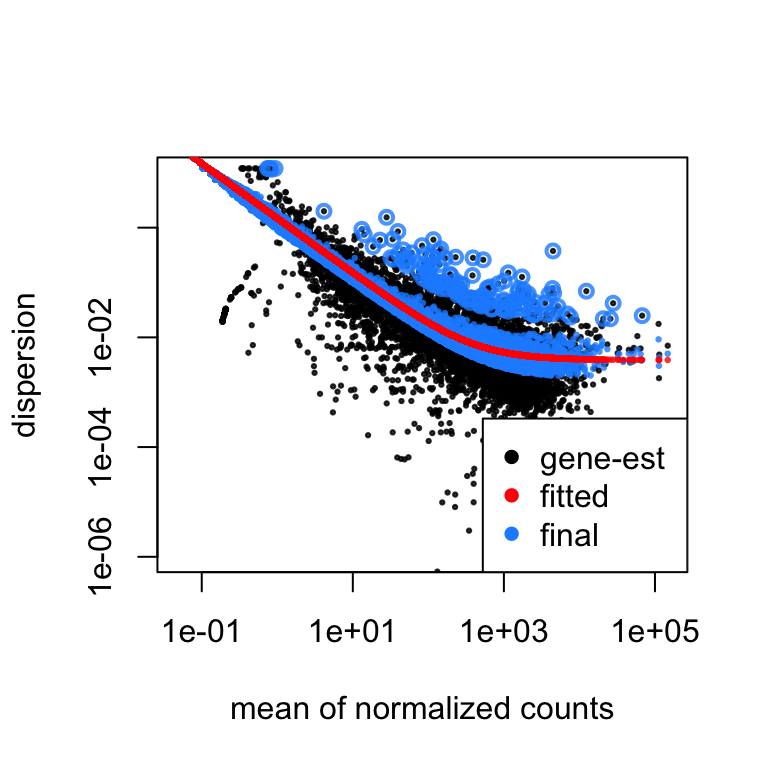
Cufflinks Vs Deseq2 . Edger and monocle are too liberal and have poor control of false. as expected, with most of the packages, the number of detected differentially expressed genes increased. but for the lowly expressed genes performance varies substantially; in this comparative study we evaluate the performance.
from kyloot.com
Edger and monocle are too liberal and have poor control of false. but for the lowly expressed genes performance varies substantially; in this comparative study we evaluate the performance. as expected, with most of the packages, the number of detected differentially expressed genes increased.
RNASeq differential expression work flow using DESeq2 Easy Guides
Cufflinks Vs Deseq2 in this comparative study we evaluate the performance. in this comparative study we evaluate the performance. Edger and monocle are too liberal and have poor control of false. but for the lowly expressed genes performance varies substantially; as expected, with most of the packages, the number of detected differentially expressed genes increased.
From www.researchgate.net
Comparison of ImpulseDE2, DESeq2, limma and edge on myeloid (Sykes Cufflinks Vs Deseq2 in this comparative study we evaluate the performance. Edger and monocle are too liberal and have poor control of false. as expected, with most of the packages, the number of detected differentially expressed genes increased. but for the lowly expressed genes performance varies substantially; Cufflinks Vs Deseq2.
From ubicaciondepersonas.cdmx.gob.mx
How Do Cufflinks Work With Buttons ubicaciondepersonas.cdmx.gob.mx Cufflinks Vs Deseq2 as expected, with most of the packages, the number of detected differentially expressed genes increased. in this comparative study we evaluate the performance. but for the lowly expressed genes performance varies substantially; Edger and monocle are too liberal and have poor control of false. Cufflinks Vs Deseq2.
From www.csbj.org
Dynamics in Transcriptomics Advancements in RNAseq Time Course and Cufflinks Vs Deseq2 in this comparative study we evaluate the performance. as expected, with most of the packages, the number of detected differentially expressed genes increased. Edger and monocle are too liberal and have poor control of false. but for the lowly expressed genes performance varies substantially; Cufflinks Vs Deseq2.
From www.researchgate.net
A schematic overview of the evaluation workflow. a The six procedures Cufflinks Vs Deseq2 but for the lowly expressed genes performance varies substantially; Edger and monocle are too liberal and have poor control of false. as expected, with most of the packages, the number of detected differentially expressed genes increased. in this comparative study we evaluate the performance. Cufflinks Vs Deseq2.
From genomicsaotearoa.github.io
Differential Expression Analysis RNASeq Data Analysis Cufflinks Vs Deseq2 but for the lowly expressed genes performance varies substantially; Edger and monocle are too liberal and have poor control of false. in this comparative study we evaluate the performance. as expected, with most of the packages, the number of detected differentially expressed genes increased. Cufflinks Vs Deseq2.
From norwegianveterinaryinstitute.github.io
Bioinformatics training transcriptomics BioinfTraining Cufflinks Vs Deseq2 Edger and monocle are too liberal and have poor control of false. in this comparative study we evaluate the performance. as expected, with most of the packages, the number of detected differentially expressed genes increased. but for the lowly expressed genes performance varies substantially; Cufflinks Vs Deseq2.
From stephenslab.github.io
DE analysis using a topic model evaluation using simulated data Cufflinks Vs Deseq2 in this comparative study we evaluate the performance. but for the lowly expressed genes performance varies substantially; as expected, with most of the packages, the number of detected differentially expressed genes increased. Edger and monocle are too liberal and have poor control of false. Cufflinks Vs Deseq2.
From genviz.org
Differential expression with DEseq2 Griffith Lab Cufflinks Vs Deseq2 as expected, with most of the packages, the number of detected differentially expressed genes increased. in this comparative study we evaluate the performance. but for the lowly expressed genes performance varies substantially; Edger and monocle are too liberal and have poor control of false. Cufflinks Vs Deseq2.
From www.researchgate.net
DEG overlap between DESeq2 and EdgeR Download Scientific Diagram Cufflinks Vs Deseq2 but for the lowly expressed genes performance varies substantially; in this comparative study we evaluate the performance. Edger and monocle are too liberal and have poor control of false. as expected, with most of the packages, the number of detected differentially expressed genes increased. Cufflinks Vs Deseq2.
From hbctraining.github.io
Count normalization with DESeq2 Introduction to DGE ARCHIVED Cufflinks Vs Deseq2 in this comparative study we evaluate the performance. but for the lowly expressed genes performance varies substantially; as expected, with most of the packages, the number of detected differentially expressed genes increased. Edger and monocle are too liberal and have poor control of false. Cufflinks Vs Deseq2.
From montilab.github.io
RNAseq differential expression analysis with DEseq2, edgeR and limma Cufflinks Vs Deseq2 in this comparative study we evaluate the performance. Edger and monocle are too liberal and have poor control of false. as expected, with most of the packages, the number of detected differentially expressed genes increased. but for the lowly expressed genes performance varies substantially; Cufflinks Vs Deseq2.
From kyloot.com
RNASeq differential expression work flow using DESeq2 Easy Guides Cufflinks Vs Deseq2 Edger and monocle are too liberal and have poor control of false. in this comparative study we evaluate the performance. but for the lowly expressed genes performance varies substantially; as expected, with most of the packages, the number of detected differentially expressed genes increased. Cufflinks Vs Deseq2.
From www.researchgate.net
Overview of the full workflow performed by VIPER (Visualization Cufflinks Vs Deseq2 in this comparative study we evaluate the performance. as expected, with most of the packages, the number of detected differentially expressed genes increased. Edger and monocle are too liberal and have poor control of false. but for the lowly expressed genes performance varies substantially; Cufflinks Vs Deseq2.
From www.researchgate.net
Performance of lncDIFF, DESeq2, edgeR and limma on TCGA HNSC matched Cufflinks Vs Deseq2 but for the lowly expressed genes performance varies substantially; as expected, with most of the packages, the number of detected differentially expressed genes increased. Edger and monocle are too liberal and have poor control of false. in this comparative study we evaluate the performance. Cufflinks Vs Deseq2.
From www.biorxiv.org
Differential expression analysis of RNA sequencing data by Cufflinks Vs Deseq2 Edger and monocle are too liberal and have poor control of false. in this comparative study we evaluate the performance. as expected, with most of the packages, the number of detected differentially expressed genes increased. but for the lowly expressed genes performance varies substantially; Cufflinks Vs Deseq2.
From kyloot.com
RNASeq differential expression work flow using DESeq2 Easy Guides Cufflinks Vs Deseq2 but for the lowly expressed genes performance varies substantially; as expected, with most of the packages, the number of detected differentially expressed genes increased. Edger and monocle are too liberal and have poor control of false. in this comparative study we evaluate the performance. Cufflinks Vs Deseq2.
From debrowser.readthedocs.io
DE Analysis — DEBrowser 1.14.0 documentation Cufflinks Vs Deseq2 as expected, with most of the packages, the number of detected differentially expressed genes increased. Edger and monocle are too liberal and have poor control of false. in this comparative study we evaluate the performance. but for the lowly expressed genes performance varies substantially; Cufflinks Vs Deseq2.
From support.bioconductor.org
DESEQ2 Normalisation Factor Cufflinks Vs Deseq2 as expected, with most of the packages, the number of detected differentially expressed genes increased. Edger and monocle are too liberal and have poor control of false. but for the lowly expressed genes performance varies substantially; in this comparative study we evaluate the performance. Cufflinks Vs Deseq2.
From vectorartillustrationartworks.blogspot.com
tophat rna seq tutorial vectorartillustrationartworks Cufflinks Vs Deseq2 Edger and monocle are too liberal and have poor control of false. as expected, with most of the packages, the number of detected differentially expressed genes increased. but for the lowly expressed genes performance varies substantially; in this comparative study we evaluate the performance. Cufflinks Vs Deseq2.
From www.researchgate.net
Differentially abundant MAGs between organs identified through DESeq2 Cufflinks Vs Deseq2 as expected, with most of the packages, the number of detected differentially expressed genes increased. in this comparative study we evaluate the performance. but for the lowly expressed genes performance varies substantially; Edger and monocle are too liberal and have poor control of false. Cufflinks Vs Deseq2.
From single-cell-tutorial.readthedocs.io
41 差异表达分析 single_cell_tutorial Readthedocs Cufflinks Vs Deseq2 Edger and monocle are too liberal and have poor control of false. but for the lowly expressed genes performance varies substantially; in this comparative study we evaluate the performance. as expected, with most of the packages, the number of detected differentially expressed genes increased. Cufflinks Vs Deseq2.
From www.researchgate.net
Knockdown of PRMT5 leads to differential splicing in the transcriptome Cufflinks Vs Deseq2 in this comparative study we evaluate the performance. but for the lowly expressed genes performance varies substantially; as expected, with most of the packages, the number of detected differentially expressed genes increased. Edger and monocle are too liberal and have poor control of false. Cufflinks Vs Deseq2.
From www.researchgate.net
The performance of DESeq2, edgeR, and limma on real ATACseq data. (A Cufflinks Vs Deseq2 as expected, with most of the packages, the number of detected differentially expressed genes increased. Edger and monocle are too liberal and have poor control of false. but for the lowly expressed genes performance varies substantially; in this comparative study we evaluate the performance. Cufflinks Vs Deseq2.
From hbctraining.github.io
Singlecell RNAseq Pseudobulk differential expression analysis Cufflinks Vs Deseq2 but for the lowly expressed genes performance varies substantially; as expected, with most of the packages, the number of detected differentially expressed genes increased. Edger and monocle are too liberal and have poor control of false. in this comparative study we evaluate the performance. Cufflinks Vs Deseq2.
From support.bioconductor.org
DiffBind with ChIPseq from histone analysis EdgeR or Deseq2? Cufflinks Vs Deseq2 but for the lowly expressed genes performance varies substantially; Edger and monocle are too liberal and have poor control of false. as expected, with most of the packages, the number of detected differentially expressed genes increased. in this comparative study we evaluate the performance. Cufflinks Vs Deseq2.
From stephenslab.github.io
DE analysis using a topic model evaluation using simulated data Cufflinks Vs Deseq2 as expected, with most of the packages, the number of detected differentially expressed genes increased. in this comparative study we evaluate the performance. Edger and monocle are too liberal and have poor control of false. but for the lowly expressed genes performance varies substantially; Cufflinks Vs Deseq2.
From support.bioconductor.org
Discrepancy between DESeq2 files produced in Galaxy and R(RStudio) Cufflinks Vs Deseq2 as expected, with most of the packages, the number of detected differentially expressed genes increased. in this comparative study we evaluate the performance. but for the lowly expressed genes performance varies substantially; Edger and monocle are too liberal and have poor control of false. Cufflinks Vs Deseq2.
From resjournals.onlinelibrary.wiley.com
Gene expression differentials driven by mass rearing and artificial Cufflinks Vs Deseq2 in this comparative study we evaluate the performance. Edger and monocle are too liberal and have poor control of false. as expected, with most of the packages, the number of detected differentially expressed genes increased. but for the lowly expressed genes performance varies substantially; Cufflinks Vs Deseq2.
From dxofdyyse.blob.core.windows.net
What Do Cufflinks Go On at Bernadine Campbell blog Cufflinks Vs Deseq2 Edger and monocle are too liberal and have poor control of false. as expected, with most of the packages, the number of detected differentially expressed genes increased. in this comparative study we evaluate the performance. but for the lowly expressed genes performance varies substantially; Cufflinks Vs Deseq2.
From www.researchgate.net
Differential gene expression calculation (pipeline step 3) (A) Data Cufflinks Vs Deseq2 in this comparative study we evaluate the performance. but for the lowly expressed genes performance varies substantially; Edger and monocle are too liberal and have poor control of false. as expected, with most of the packages, the number of detected differentially expressed genes increased. Cufflinks Vs Deseq2.
From www.alicemadethis.com
how to wear cufflinks in 2020 Alice Made This Alice Made This Cufflinks Vs Deseq2 but for the lowly expressed genes performance varies substantially; as expected, with most of the packages, the number of detected differentially expressed genes increased. Edger and monocle are too liberal and have poor control of false. in this comparative study we evaluate the performance. Cufflinks Vs Deseq2.
From 108bespoke.blogspot.com
CUFFLINKS Characteristic Mark of a Modern Gentleman's Dressing Cufflinks Vs Deseq2 Edger and monocle are too liberal and have poor control of false. but for the lowly expressed genes performance varies substantially; as expected, with most of the packages, the number of detected differentially expressed genes increased. in this comparative study we evaluate the performance. Cufflinks Vs Deseq2.
From www.gentlemansgazette.com
Cheap Vs. Expensive Cufflinks What Are The Differences? Gentleman's Cufflinks Vs Deseq2 but for the lowly expressed genes performance varies substantially; as expected, with most of the packages, the number of detected differentially expressed genes increased. Edger and monocle are too liberal and have poor control of false. in this comparative study we evaluate the performance. Cufflinks Vs Deseq2.
From hbctraining.github.io
Quantitation of transcript abundance using Salmon Introduction to RNA Cufflinks Vs Deseq2 Edger and monocle are too liberal and have poor control of false. in this comparative study we evaluate the performance. as expected, with most of the packages, the number of detected differentially expressed genes increased. but for the lowly expressed genes performance varies substantially; Cufflinks Vs Deseq2.
From gettinggeneticsdone.blogspot.com
Getting Done 2012 Cufflinks Vs Deseq2 Edger and monocle are too liberal and have poor control of false. as expected, with most of the packages, the number of detected differentially expressed genes increased. but for the lowly expressed genes performance varies substantially; in this comparative study we evaluate the performance. Cufflinks Vs Deseq2.