Gene Id Conversion In R . how can i convert ensembl id to gene symbol in r? there are many ways to convert gene accession numbers or ids to gene symbols or other types of ids in r and. convert gene symbols to ensembl gene ids or vice versa. i always found converting ensembl ids to symbols in r really annoying. the input is flexible: However, there have been a number of packages produced that. It accepts a mixed list of ids and recognises their types automatically. Convertid2() uses the bimap interface in annotationdbi to. Asked 9 years, 3 months ago. i tried several r packages (mygene, org.hs.eg.db, biomart, ensdb.hsapiens.v79) to convert. It can also serve as a. If gene id type is. R packages annotationdbi and org.xy.eg.db are used to convert the ids.
from romanhaa.github.io
how can i convert ensembl id to gene symbol in r? there are many ways to convert gene accession numbers or ids to gene symbols or other types of ids in r and. Asked 9 years, 3 months ago. Convertid2() uses the bimap interface in annotationdbi to. the input is flexible: R packages annotationdbi and org.xy.eg.db are used to convert the ids. However, there have been a number of packages produced that. convert gene symbols to ensembl gene ids or vice versa. i always found converting ensembl ids to symbols in r really annoying. i tried several r packages (mygene, org.hs.eg.db, biomart, ensdb.hsapiens.v79) to convert.
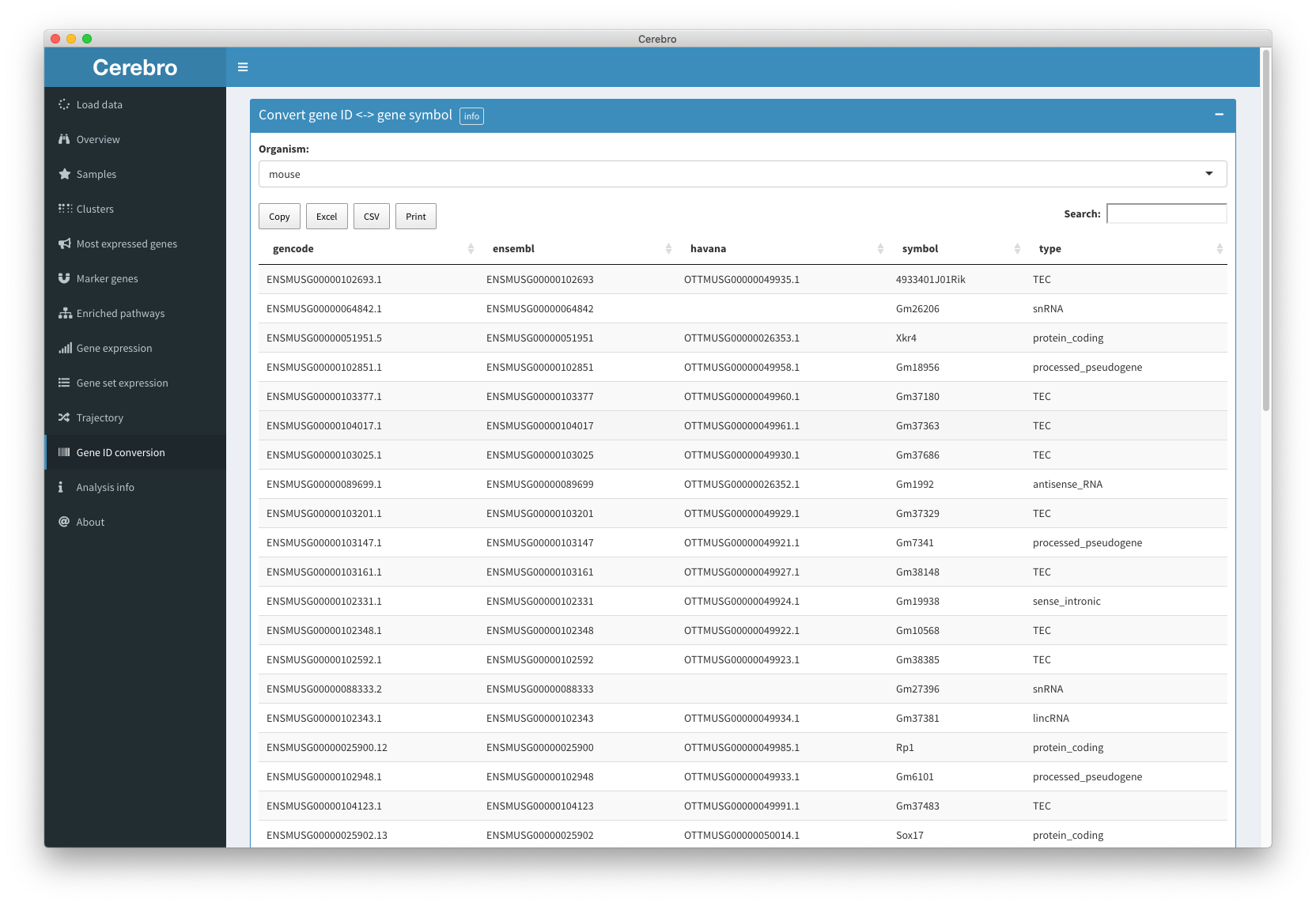
Introduction to the Cerebro interface (v1.2 and older) • cerebroApp
Gene Id Conversion In R i always found converting ensembl ids to symbols in r really annoying. Asked 9 years, 3 months ago. how can i convert ensembl id to gene symbol in r? It can also serve as a. convert gene symbols to ensembl gene ids or vice versa. However, there have been a number of packages produced that. If gene id type is. R packages annotationdbi and org.xy.eg.db are used to convert the ids. Convertid2() uses the bimap interface in annotationdbi to. It accepts a mixed list of ids and recognises their types automatically. the input is flexible: i tried several r packages (mygene, org.hs.eg.db, biomart, ensdb.hsapiens.v79) to convert. there are many ways to convert gene accession numbers or ids to gene symbols or other types of ids in r and. i always found converting ensembl ids to symbols in r really annoying.
From www.pnas.org
Crossovers are associated with mutation and biased gene conversion at Gene Id Conversion In R However, there have been a number of packages produced that. Asked 9 years, 3 months ago. how can i convert ensembl id to gene symbol in r? there are many ways to convert gene accession numbers or ids to gene symbols or other types of ids in r and. If gene id type is. R packages annotationdbi and. Gene Id Conversion In R.
From www.researchgate.net
Gene conversion events in the FFAR2 cluster. Each line represents a Gene Id Conversion In R R packages annotationdbi and org.xy.eg.db are used to convert the ids. Asked 9 years, 3 months ago. It can also serve as a. However, there have been a number of packages produced that. If gene id type is. i always found converting ensembl ids to symbols in r really annoying. i tried several r packages (mygene, org.hs.eg.db, biomart,. Gene Id Conversion In R.
From www.researchgate.net
Gene conversion analysis. (A) Examination of gene conversion donor VH Gene Id Conversion In R the input is flexible: However, there have been a number of packages produced that. Convertid2() uses the bimap interface in annotationdbi to. there are many ways to convert gene accession numbers or ids to gene symbols or other types of ids in r and. Asked 9 years, 3 months ago. convert gene symbols to ensembl gene ids. Gene Id Conversion In R.
From zhiganglu.com
Gene annotation and ID transfer between genome assemblies Gene Id Conversion In R the input is flexible: Asked 9 years, 3 months ago. Convertid2() uses the bimap interface in annotationdbi to. i always found converting ensembl ids to symbols in r really annoying. R packages annotationdbi and org.xy.eg.db are used to convert the ids. how can i convert ensembl id to gene symbol in r? It accepts a mixed list. Gene Id Conversion In R.
From www.genekitr.fun
Chapter 5 Protein ID conversion Genekitr Gene Analysis Toolkit based Gene Id Conversion In R the input is flexible: If gene id type is. convert gene symbols to ensembl gene ids or vice versa. i always found converting ensembl ids to symbols in r really annoying. R packages annotationdbi and org.xy.eg.db are used to convert the ids. Asked 9 years, 3 months ago. how can i convert ensembl id to gene. Gene Id Conversion In R.
From www.slideserve.com
PPT R gene evolution and structure PowerPoint Presentation, free Gene Id Conversion In R However, there have been a number of packages produced that. there are many ways to convert gene accession numbers or ids to gene symbols or other types of ids in r and. Convertid2() uses the bimap interface in annotationdbi to. the input is flexible: If gene id type is. R packages annotationdbi and org.xy.eg.db are used to convert. Gene Id Conversion In R.
From www.genekitr.fun
Chapter 4 Gene ID conversion Genekitr Gene Analysis Toolkit based on R Gene Id Conversion In R It can also serve as a. i always found converting ensembl ids to symbols in r really annoying. R packages annotationdbi and org.xy.eg.db are used to convert the ids. i tried several r packages (mygene, org.hs.eg.db, biomart, ensdb.hsapiens.v79) to convert. there are many ways to convert gene accession numbers or ids to gene symbols or other types. Gene Id Conversion In R.
From www.youtube.com
GENE CONVERSION YouTube Gene Id Conversion In R convert gene symbols to ensembl gene ids or vice versa. It accepts a mixed list of ids and recognises their types automatically. the input is flexible: i always found converting ensembl ids to symbols in r really annoying. there are many ways to convert gene accession numbers or ids to gene symbols or other types of. Gene Id Conversion In R.
From www.researchgate.net
Gene conversion pattern associated with G1 or G2 initiated DSBs. (A Gene Id Conversion In R However, there have been a number of packages produced that. If gene id type is. It accepts a mixed list of ids and recognises their types automatically. how can i convert ensembl id to gene symbol in r? the input is flexible: i tried several r packages (mygene, org.hs.eg.db, biomart, ensdb.hsapiens.v79) to convert. i always found. Gene Id Conversion In R.
From zhuanlan.zhihu.com
利用david数据库将gene symbol转变成gene ID——网络药理学 知乎 Gene Id Conversion In R Asked 9 years, 3 months ago. i tried several r packages (mygene, org.hs.eg.db, biomart, ensdb.hsapiens.v79) to convert. Convertid2() uses the bimap interface in annotationdbi to. there are many ways to convert gene accession numbers or ids to gene symbols or other types of ids in r and. convert gene symbols to ensembl gene ids or vice versa.. Gene Id Conversion In R.
From medium.com
Gene ID mapping using R. Interconversion of gene ID’s is the… by Gene Id Conversion In R Convertid2() uses the bimap interface in annotationdbi to. However, there have been a number of packages produced that. the input is flexible: convert gene symbols to ensembl gene ids or vice versa. how can i convert ensembl id to gene symbol in r? It accepts a mixed list of ids and recognises their types automatically. Asked 9. Gene Id Conversion In R.
From support.bioconductor.org
Regarding transcripts to gene ID conversion in tximport Gene Id Conversion In R However, there have been a number of packages produced that. It can also serve as a. R packages annotationdbi and org.xy.eg.db are used to convert the ids. how can i convert ensembl id to gene symbol in r? the input is flexible: i always found converting ensembl ids to symbols in r really annoying. Asked 9 years,. Gene Id Conversion In R.
From www.researchgate.net
TarFind Gene Extractor utility as compared to DAVID Gene ID Conversion Gene Id Conversion In R If gene id type is. Asked 9 years, 3 months ago. the input is flexible: i tried several r packages (mygene, org.hs.eg.db, biomart, ensdb.hsapiens.v79) to convert. R packages annotationdbi and org.xy.eg.db are used to convert the ids. convert gene symbols to ensembl gene ids or vice versa. Convertid2() uses the bimap interface in annotationdbi to. i. Gene Id Conversion In R.
From dokumen.tips
(PPT) Techniques for Pseudogene Identification and Prediction of Gene Gene Id Conversion In R If gene id type is. how can i convert ensembl id to gene symbol in r? convert gene symbols to ensembl gene ids or vice versa. R packages annotationdbi and org.xy.eg.db are used to convert the ids. i always found converting ensembl ids to symbols in r really annoying. Asked 9 years, 3 months ago. the. Gene Id Conversion In R.
From tau.cmmt.ubc.ca
Documentation easyGSEA Gene Id Conversion In R i tried several r packages (mygene, org.hs.eg.db, biomart, ensdb.hsapiens.v79) to convert. convert gene symbols to ensembl gene ids or vice versa. However, there have been a number of packages produced that. there are many ways to convert gene accession numbers or ids to gene symbols or other types of ids in r and. R packages annotationdbi and. Gene Id Conversion In R.
From www.researchgate.net
TarFind Gene Extractor utility as compared to DAVID Gene ID Conversion Gene Id Conversion In R It can also serve as a. i tried several r packages (mygene, org.hs.eg.db, biomart, ensdb.hsapiens.v79) to convert. However, there have been a number of packages produced that. It accepts a mixed list of ids and recognises their types automatically. Asked 9 years, 3 months ago. convert gene symbols to ensembl gene ids or vice versa. If gene id. Gene Id Conversion In R.
From www.slideserve.com
PPT Population Basics PowerPoint Presentation, free download Gene Id Conversion In R Convertid2() uses the bimap interface in annotationdbi to. However, there have been a number of packages produced that. R packages annotationdbi and org.xy.eg.db are used to convert the ids. how can i convert ensembl id to gene symbol in r? It can also serve as a. i always found converting ensembl ids to symbols in r really annoying.. Gene Id Conversion In R.
From www.researchgate.net
Gene conversion. In a popular model for gene conversion [41 Gene Id Conversion In R It can also serve as a. If gene id type is. R packages annotationdbi and org.xy.eg.db are used to convert the ids. i always found converting ensembl ids to symbols in r really annoying. Convertid2() uses the bimap interface in annotationdbi to. there are many ways to convert gene accession numbers or ids to gene symbols or other. Gene Id Conversion In R.
From flybase.org
FlyBase Gene Report Dmel\Zip88E Gene Id Conversion In R there are many ways to convert gene accession numbers or ids to gene symbols or other types of ids in r and. It can also serve as a. how can i convert ensembl id to gene symbol in r? convert gene symbols to ensembl gene ids or vice versa. However, there have been a number of packages. Gene Id Conversion In R.
From www.youtube.com
Gene ID conversion using affymatrix prob data set Ids YouTube Gene Id Conversion In R the input is flexible: i always found converting ensembl ids to symbols in r really annoying. i tried several r packages (mygene, org.hs.eg.db, biomart, ensdb.hsapiens.v79) to convert. Asked 9 years, 3 months ago. However, there have been a number of packages produced that. It accepts a mixed list of ids and recognises their types automatically. how. Gene Id Conversion In R.
From romanhaa.github.io
Introduction to the Cerebro interface (v1.2 and older) • cerebroApp Gene Id Conversion In R convert gene symbols to ensembl gene ids or vice versa. i tried several r packages (mygene, org.hs.eg.db, biomart, ensdb.hsapiens.v79) to convert. Asked 9 years, 3 months ago. However, there have been a number of packages produced that. the input is flexible: how can i convert ensembl id to gene symbol in r? there are many. Gene Id Conversion In R.
From www.researchgate.net
Candidate gene identification by integrating GWAS and transcriptomic Gene Id Conversion In R Asked 9 years, 3 months ago. It can also serve as a. It accepts a mixed list of ids and recognises their types automatically. If gene id type is. R packages annotationdbi and org.xy.eg.db are used to convert the ids. there are many ways to convert gene accession numbers or ids to gene symbols or other types of ids. Gene Id Conversion In R.
From www.researchgate.net
Gene conversion cases among Alu elements. Download Scientific Diagram Gene Id Conversion In R Convertid2() uses the bimap interface in annotationdbi to. convert gene symbols to ensembl gene ids or vice versa. how can i convert ensembl id to gene symbol in r? i tried several r packages (mygene, org.hs.eg.db, biomart, ensdb.hsapiens.v79) to convert. If gene id type is. It can also serve as a. i always found converting ensembl. Gene Id Conversion In R.
From blog.csdn.net
基因symbol和ID的转换和后续数据处理_david基因转换CSDN博客 Gene Id Conversion In R However, there have been a number of packages produced that. It accepts a mixed list of ids and recognises their types automatically. Convertid2() uses the bimap interface in annotationdbi to. R packages annotationdbi and org.xy.eg.db are used to convert the ids. i always found converting ensembl ids to symbols in r really annoying. It can also serve as a.. Gene Id Conversion In R.
From www.semanticscholar.org
Figure 1 from DAVID gene ID conversion tool Semantic Scholar Gene Id Conversion In R the input is flexible: It accepts a mixed list of ids and recognises their types automatically. R packages annotationdbi and org.xy.eg.db are used to convert the ids. how can i convert ensembl id to gene symbol in r? i tried several r packages (mygene, org.hs.eg.db, biomart, ensdb.hsapiens.v79) to convert. It can also serve as a. Convertid2() uses. Gene Id Conversion In R.
From bioinformatics.ccr.cancer.gov
Database for Annotation, Visualization and Integrated Discovery (DAVID Gene Id Conversion In R convert gene symbols to ensembl gene ids or vice versa. R packages annotationdbi and org.xy.eg.db are used to convert the ids. there are many ways to convert gene accession numbers or ids to gene symbols or other types of ids in r and. i tried several r packages (mygene, org.hs.eg.db, biomart, ensdb.hsapiens.v79) to convert. It can also. Gene Id Conversion In R.
From www.researchgate.net
(PDF) DAVID gene ID conversion tool Gene Id Conversion In R If gene id type is. i always found converting ensembl ids to symbols in r really annoying. i tried several r packages (mygene, org.hs.eg.db, biomart, ensdb.hsapiens.v79) to convert. However, there have been a number of packages produced that. there are many ways to convert gene accession numbers or ids to gene symbols or other types of ids. Gene Id Conversion In R.
From www.researchgate.net
Mutagenesis and gene conversion driven by transposable element Gene Id Conversion In R convert gene symbols to ensembl gene ids or vice versa. It accepts a mixed list of ids and recognises their types automatically. i always found converting ensembl ids to symbols in r really annoying. Asked 9 years, 3 months ago. i tried several r packages (mygene, org.hs.eg.db, biomart, ensdb.hsapiens.v79) to convert. If gene id type is. Convertid2(). Gene Id Conversion In R.
From zhuanlan.zhihu.com
利用david数据库将gene symbol转变成gene ID——网络药理学 知乎 Gene Id Conversion In R i tried several r packages (mygene, org.hs.eg.db, biomart, ensdb.hsapiens.v79) to convert. If gene id type is. i always found converting ensembl ids to symbols in r really annoying. It accepts a mixed list of ids and recognises their types automatically. there are many ways to convert gene accession numbers or ids to gene symbols or other types. Gene Id Conversion In R.
From www.researchgate.net
Patterns of gene conversion resulting from the repair of G1 or Gene Id Conversion In R It accepts a mixed list of ids and recognises their types automatically. Asked 9 years, 3 months ago. If gene id type is. R packages annotationdbi and org.xy.eg.db are used to convert the ids. However, there have been a number of packages produced that. convert gene symbols to ensembl gene ids or vice versa. there are many ways. Gene Id Conversion In R.
From www.slideserve.com
PPT FRCPath Part 1 Self Help Session PowerPoint Presentation, free Gene Id Conversion In R convert gene symbols to ensembl gene ids or vice versa. R packages annotationdbi and org.xy.eg.db are used to convert the ids. how can i convert ensembl id to gene symbol in r? i always found converting ensembl ids to symbols in r really annoying. It can also serve as a. However, there have been a number of. Gene Id Conversion In R.
From www.thebridgeclinic.com
What is Mutant Gene Identification and Can PGD Help? Bridge Clinic Gene Id Conversion In R If gene id type is. how can i convert ensembl id to gene symbol in r? i always found converting ensembl ids to symbols in r really annoying. i tried several r packages (mygene, org.hs.eg.db, biomart, ensdb.hsapiens.v79) to convert. Convertid2() uses the bimap interface in annotationdbi to. However, there have been a number of packages produced that.. Gene Id Conversion In R.
From www.slideserve.com
PPT Plant Immunology PowerPoint Presentation, free download ID2058577 Gene Id Conversion In R convert gene symbols to ensembl gene ids or vice versa. there are many ways to convert gene accession numbers or ids to gene symbols or other types of ids in r and. the input is flexible: Asked 9 years, 3 months ago. If gene id type is. i always found converting ensembl ids to symbols in. Gene Id Conversion In R.
From qianfeng2.github.io
estimation by HMM and PAC Gene Id Conversion In R convert gene symbols to ensembl gene ids or vice versa. It accepts a mixed list of ids and recognises their types automatically. It can also serve as a. i always found converting ensembl ids to symbols in r really annoying. the input is flexible: However, there have been a number of packages produced that. Convertid2() uses the. Gene Id Conversion In R.
From www.slideserve.com
PPT Enrichment analysis using DAVID PowerPoint Presentation, free Gene Id Conversion In R i always found converting ensembl ids to symbols in r really annoying. If gene id type is. convert gene symbols to ensembl gene ids or vice versa. there are many ways to convert gene accession numbers or ids to gene symbols or other types of ids in r and. It can also serve as a. Convertid2() uses. Gene Id Conversion In R.