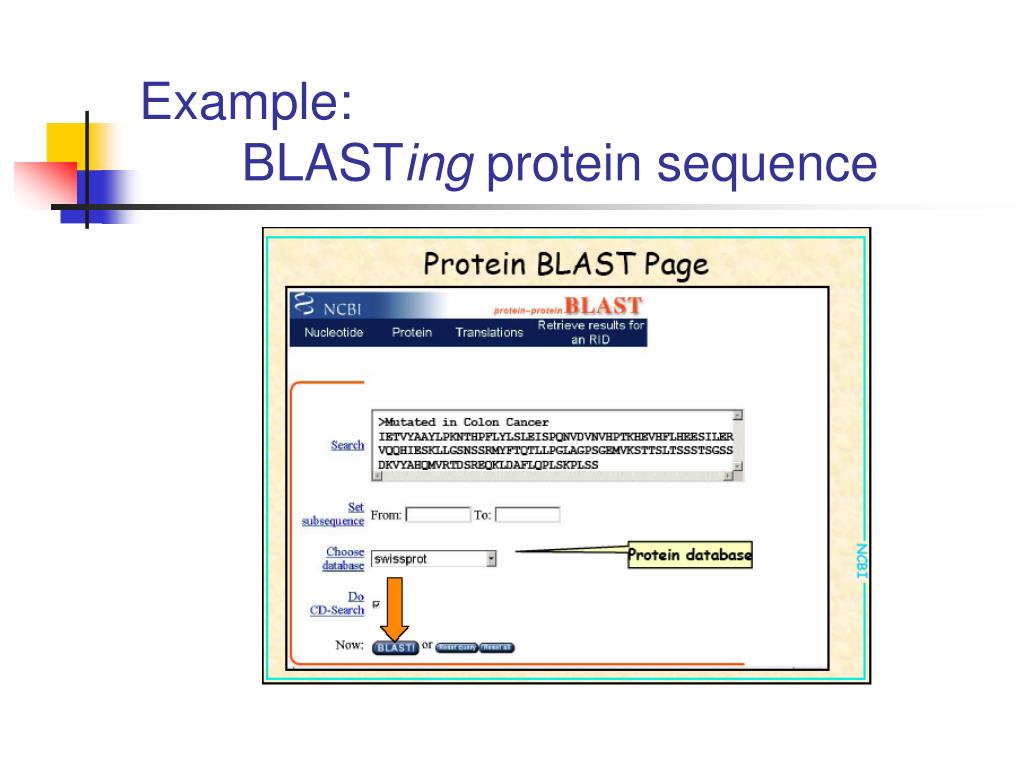
How Does Protein Blast Work . blastp (protein blast): Blastp, or protein blast, is used to compare protein. The blast computers start with a small set of. blastp simply compares a protein query to a protein database. the basic local alignment search tool (blast) finds regions of local similarity between sequences. The program compares nucleotide or protein. the basic local alignment search tool (blast) finds regions of local similarity between protein or nucleotide sequences. protein sequence being compared against nr database using blastp. in this kind of blast, the query sequence (dna / protein) is searched against an existing collection of conserved. Compares one or more protein query sequences to a subject protein sequence or a database of protein. blast works by detecting local alignments between sequences that work the best.
from www.slideserve.com
The blast computers start with a small set of. the basic local alignment search tool (blast) finds regions of local similarity between sequences. in this kind of blast, the query sequence (dna / protein) is searched against an existing collection of conserved. the basic local alignment search tool (blast) finds regions of local similarity between protein or nucleotide sequences. blast works by detecting local alignments between sequences that work the best. Compares one or more protein query sequences to a subject protein sequence or a database of protein. protein sequence being compared against nr database using blastp. Blastp, or protein blast, is used to compare protein. The program compares nucleotide or protein. blastp simply compares a protein query to a protein database.
PPT NCBI PowerPoint Presentation ID315849
How Does Protein Blast Work Blastp, or protein blast, is used to compare protein. blastp simply compares a protein query to a protein database. Blastp, or protein blast, is used to compare protein. blast works by detecting local alignments between sequences that work the best. in this kind of blast, the query sequence (dna / protein) is searched against an existing collection of conserved. The program compares nucleotide or protein. the basic local alignment search tool (blast) finds regions of local similarity between sequences. blastp (protein blast): protein sequence being compared against nr database using blastp. Compares one or more protein query sequences to a subject protein sequence or a database of protein. the basic local alignment search tool (blast) finds regions of local similarity between protein or nucleotide sequences. The blast computers start with a small set of.
From www.youtube.com
How to Use BLAST for Finding and Aligning DNA or Protein Sequences How Does Protein Blast Work blastp (protein blast): blast works by detecting local alignments between sequences that work the best. The program compares nucleotide or protein. Blastp, or protein blast, is used to compare protein. protein sequence being compared against nr database using blastp. The blast computers start with a small set of. in this kind of blast, the query sequence. How Does Protein Blast Work.
From www.slideserve.com
PPT BLAST PowerPoint Presentation, free download ID5789334 How Does Protein Blast Work The program compares nucleotide or protein. the basic local alignment search tool (blast) finds regions of local similarity between protein or nucleotide sequences. blastp simply compares a protein query to a protein database. the basic local alignment search tool (blast) finds regions of local similarity between sequences. Blastp, or protein blast, is used to compare protein. The. How Does Protein Blast Work.
From dokumen.tips
(PDF) Introduction to BLAST with Protein Sequencesmath.usu.edu How Does Protein Blast Work blastp (protein blast): Compares one or more protein query sequences to a subject protein sequence or a database of protein. The program compares nucleotide or protein. the basic local alignment search tool (blast) finds regions of local similarity between sequences. Blastp, or protein blast, is used to compare protein. the basic local alignment search tool (blast) finds. How Does Protein Blast Work.
From www.slideserve.com
PPT Diversity of Life Introduction to Biological Classification How Does Protein Blast Work protein sequence being compared against nr database using blastp. the basic local alignment search tool (blast) finds regions of local similarity between sequences. in this kind of blast, the query sequence (dna / protein) is searched against an existing collection of conserved. blastp (protein blast): The program compares nucleotide or protein. Compares one or more protein. How Does Protein Blast Work.
From www.youtube.com
Which Database to Choose for Protein BLAST run YouTube How Does Protein Blast Work blastp simply compares a protein query to a protein database. the basic local alignment search tool (blast) finds regions of local similarity between protein or nucleotide sequences. Compares one or more protein query sequences to a subject protein sequence or a database of protein. Blastp, or protein blast, is used to compare protein. blast works by detecting. How Does Protein Blast Work.
From www.slideserve.com
PPT Introduction to BLAST PowerPoint Presentation, free download ID How Does Protein Blast Work in this kind of blast, the query sequence (dna / protein) is searched against an existing collection of conserved. The blast computers start with a small set of. blastp simply compares a protein query to a protein database. Blastp, or protein blast, is used to compare protein. The program compares nucleotide or protein. Compares one or more protein. How Does Protein Blast Work.
From www.slideserve.com
PPT BLAST PowerPoint Presentation, free download ID4734821 How Does Protein Blast Work Compares one or more protein query sequences to a subject protein sequence or a database of protein. Blastp, or protein blast, is used to compare protein. The blast computers start with a small set of. The program compares nucleotide or protein. the basic local alignment search tool (blast) finds regions of local similarity between sequences. blast works by. How Does Protein Blast Work.
From www.slideserve.com
PPT Introduction to BLAST PowerPoint Presentation, free download ID How Does Protein Blast Work The program compares nucleotide or protein. in this kind of blast, the query sequence (dna / protein) is searched against an existing collection of conserved. Blastp, or protein blast, is used to compare protein. blastp (protein blast): the basic local alignment search tool (blast) finds regions of local similarity between protein or nucleotide sequences. protein sequence. How Does Protein Blast Work.
From www.researchgate.net
(PDF) Is Protein BLAST a thing of the past? How Does Protein Blast Work the basic local alignment search tool (blast) finds regions of local similarity between sequences. protein sequence being compared against nr database using blastp. blastp (protein blast): Blastp, or protein blast, is used to compare protein. The blast computers start with a small set of. The program compares nucleotide or protein. Compares one or more protein query sequences. How Does Protein Blast Work.
From www.youtube.com
NCBI BLAST tutorial how to use blast for finding and aligning DNA or How Does Protein Blast Work Blastp, or protein blast, is used to compare protein. Compares one or more protein query sequences to a subject protein sequence or a database of protein. protein sequence being compared against nr database using blastp. blastp (protein blast): blast works by detecting local alignments between sequences that work the best. The program compares nucleotide or protein. . How Does Protein Blast Work.
From www.researchgate.net
Protein blast similarity and secondary structure details of selected How Does Protein Blast Work in this kind of blast, the query sequence (dna / protein) is searched against an existing collection of conserved. blast works by detecting local alignments between sequences that work the best. the basic local alignment search tool (blast) finds regions of local similarity between sequences. the basic local alignment search tool (blast) finds regions of local. How Does Protein Blast Work.
From www.researchgate.net
NCBI Protein BLAST Taxonomy OmpC321340 Download Scientific Diagram How Does Protein Blast Work protein sequence being compared against nr database using blastp. the basic local alignment search tool (blast) finds regions of local similarity between sequences. The program compares nucleotide or protein. blastp (protein blast): Compares one or more protein query sequences to a subject protein sequence or a database of protein. The blast computers start with a small set. How Does Protein Blast Work.
From mammothmemory.net
Example of how protease enzymes break down proteins How Does Protein Blast Work the basic local alignment search tool (blast) finds regions of local similarity between protein or nucleotide sequences. the basic local alignment search tool (blast) finds regions of local similarity between sequences. Blastp, or protein blast, is used to compare protein. The program compares nucleotide or protein. Compares one or more protein query sequences to a subject protein sequence. How Does Protein Blast Work.
From www.slideserve.com
PPT BLAST PowerPoint Presentation, free download ID3384708 How Does Protein Blast Work the basic local alignment search tool (blast) finds regions of local similarity between sequences. The blast computers start with a small set of. blastp simply compares a protein query to a protein database. The program compares nucleotide or protein. blastp (protein blast): in this kind of blast, the query sequence (dna / protein) is searched against. How Does Protein Blast Work.
From www.youtube.com
Protein to Translated Nucleotide BLAST tBLASTn YouTube How Does Protein Blast Work The program compares nucleotide or protein. protein sequence being compared against nr database using blastp. the basic local alignment search tool (blast) finds regions of local similarity between protein or nucleotide sequences. Blastp, or protein blast, is used to compare protein. The blast computers start with a small set of. blastp simply compares a protein query to. How Does Protein Blast Work.
From www.youtube.com
Translated Nucleotide to Protein BLAST BLASTX YouTube How Does Protein Blast Work blast works by detecting local alignments between sequences that work the best. blastp (protein blast): in this kind of blast, the query sequence (dna / protein) is searched against an existing collection of conserved. the basic local alignment search tool (blast) finds regions of local similarity between sequences. the basic local alignment search tool (blast). How Does Protein Blast Work.
From www.youtube.com
BLAST Protein YouTube How Does Protein Blast Work the basic local alignment search tool (blast) finds regions of local similarity between protein or nucleotide sequences. blastp simply compares a protein query to a protein database. The blast computers start with a small set of. protein sequence being compared against nr database using blastp. Compares one or more protein query sequences to a subject protein sequence. How Does Protein Blast Work.
From www.slideserve.com
PPT NCBI PowerPoint Presentation ID315849 How Does Protein Blast Work protein sequence being compared against nr database using blastp. in this kind of blast, the query sequence (dna / protein) is searched against an existing collection of conserved. blast works by detecting local alignments between sequences that work the best. The blast computers start with a small set of. the basic local alignment search tool (blast). How Does Protein Blast Work.
From www.psb.ugent.be
PTM Viewer How Does Protein Blast Work blast works by detecting local alignments between sequences that work the best. the basic local alignment search tool (blast) finds regions of local similarity between sequences. The blast computers start with a small set of. Compares one or more protein query sequences to a subject protein sequence or a database of protein. in this kind of blast,. How Does Protein Blast Work.
From www.slideserve.com
PPT Similarity Searches on Sequence Databases PowerPoint Presentation How Does Protein Blast Work protein sequence being compared against nr database using blastp. the basic local alignment search tool (blast) finds regions of local similarity between protein or nucleotide sequences. Compares one or more protein query sequences to a subject protein sequence or a database of protein. blastp simply compares a protein query to a protein database. The blast computers start. How Does Protein Blast Work.
From www.researchgate.net
The main framework of HBLAST and the workflow on the protein sequence How Does Protein Blast Work blast works by detecting local alignments between sequences that work the best. protein sequence being compared against nr database using blastp. the basic local alignment search tool (blast) finds regions of local similarity between sequences. The program compares nucleotide or protein. blastp (protein blast): the basic local alignment search tool (blast) finds regions of local. How Does Protein Blast Work.
From www.youtube.com
bioinformatics step by step guide how to use protein blast in NCBI How Does Protein Blast Work Blastp, or protein blast, is used to compare protein. blastp simply compares a protein query to a protein database. the basic local alignment search tool (blast) finds regions of local similarity between protein or nucleotide sequences. blast works by detecting local alignments between sequences that work the best. Compares one or more protein query sequences to a. How Does Protein Blast Work.
From thebiologynotes.com
BLAST (Bioinformatics) Definition, 5 Types, Steps, Uses How Does Protein Blast Work The blast computers start with a small set of. protein sequence being compared against nr database using blastp. blastp simply compares a protein query to a protein database. the basic local alignment search tool (blast) finds regions of local similarity between protein or nucleotide sequences. the basic local alignment search tool (blast) finds regions of local. How Does Protein Blast Work.
From www.youtube.com
How Can I Use Protein BLAST (BLASTp) for My Research? YouTube How Does Protein Blast Work Compares one or more protein query sequences to a subject protein sequence or a database of protein. the basic local alignment search tool (blast) finds regions of local similarity between protein or nucleotide sequences. The blast computers start with a small set of. protein sequence being compared against nr database using blastp. blastp (protein blast): in. How Does Protein Blast Work.
From www.youtube.com
NCBI Blast Tutorial YouTube How Does Protein Blast Work the basic local alignment search tool (blast) finds regions of local similarity between protein or nucleotide sequences. in this kind of blast, the query sequence (dna / protein) is searched against an existing collection of conserved. The blast computers start with a small set of. blast works by detecting local alignments between sequences that work the best.. How Does Protein Blast Work.
From www.slideserve.com
PPT Protein Sequence PowerPoint Presentation, free download ID4271810 How Does Protein Blast Work blast works by detecting local alignments between sequences that work the best. The blast computers start with a small set of. The program compares nucleotide or protein. in this kind of blast, the query sequence (dna / protein) is searched against an existing collection of conserved. blastp (protein blast): blastp simply compares a protein query to. How Does Protein Blast Work.
From www.slideserve.com
PPT Pairwise alignment 2 Scoring matrices and gaps BLAST, BLAT and How Does Protein Blast Work blast works by detecting local alignments between sequences that work the best. in this kind of blast, the query sequence (dna / protein) is searched against an existing collection of conserved. protein sequence being compared against nr database using blastp. Blastp, or protein blast, is used to compare protein. the basic local alignment search tool (blast). How Does Protein Blast Work.
From exogcclws.blob.core.windows.net
Human Protein Blast at Antonia Stillwell blog How Does Protein Blast Work the basic local alignment search tool (blast) finds regions of local similarity between protein or nucleotide sequences. protein sequence being compared against nr database using blastp. Blastp, or protein blast, is used to compare protein. Compares one or more protein query sequences to a subject protein sequence or a database of protein. in this kind of blast,. How Does Protein Blast Work.
From widdowquinn.github.io
01blast_at_NCBI_website How Does Protein Blast Work Compares one or more protein query sequences to a subject protein sequence or a database of protein. The program compares nucleotide or protein. in this kind of blast, the query sequence (dna / protein) is searched against an existing collection of conserved. the basic local alignment search tool (blast) finds regions of local similarity between protein or nucleotide. How Does Protein Blast Work.
From abacus.bates.edu
BLASTP Results Part 1 How Does Protein Blast Work blastp (protein blast): in this kind of blast, the query sequence (dna / protein) is searched against an existing collection of conserved. blast works by detecting local alignments between sequences that work the best. the basic local alignment search tool (blast) finds regions of local similarity between protein or nucleotide sequences. The program compares nucleotide or. How Does Protein Blast Work.
From slideplayer.com
to Introduction to Bioinformatics ppt download How Does Protein Blast Work the basic local alignment search tool (blast) finds regions of local similarity between protein or nucleotide sequences. The program compares nucleotide or protein. blastp simply compares a protein query to a protein database. the basic local alignment search tool (blast) finds regions of local similarity between sequences. in this kind of blast, the query sequence (dna. How Does Protein Blast Work.
From www.slideserve.com
PPT BLAST PowerPoint Presentation, free download ID1872123 How Does Protein Blast Work the basic local alignment search tool (blast) finds regions of local similarity between protein or nucleotide sequences. the basic local alignment search tool (blast) finds regions of local similarity between sequences. Compares one or more protein query sequences to a subject protein sequence or a database of protein. protein sequence being compared against nr database using blastp.. How Does Protein Blast Work.
From www.slideserve.com
PPT Sequence Similarity & Alignment with BLAST PowerPoint How Does Protein Blast Work blastp simply compares a protein query to a protein database. the basic local alignment search tool (blast) finds regions of local similarity between protein or nucleotide sequences. blastp (protein blast): The blast computers start with a small set of. Compares one or more protein query sequences to a subject protein sequence or a database of protein. . How Does Protein Blast Work.
From www.researchgate.net
Heatmap representation of protein BLAST showing sequence divergence How Does Protein Blast Work blast works by detecting local alignments between sequences that work the best. the basic local alignment search tool (blast) finds regions of local similarity between sequences. The program compares nucleotide or protein. in this kind of blast, the query sequence (dna / protein) is searched against an existing collection of conserved. blastp simply compares a protein. How Does Protein Blast Work.
From www.youtube.com
Which BLAST Algorithm to use for ProteinProtein BLAST YouTube How Does Protein Blast Work The program compares nucleotide or protein. blast works by detecting local alignments between sequences that work the best. Blastp, or protein blast, is used to compare protein. in this kind of blast, the query sequence (dna / protein) is searched against an existing collection of conserved. the basic local alignment search tool (blast) finds regions of local. How Does Protein Blast Work.