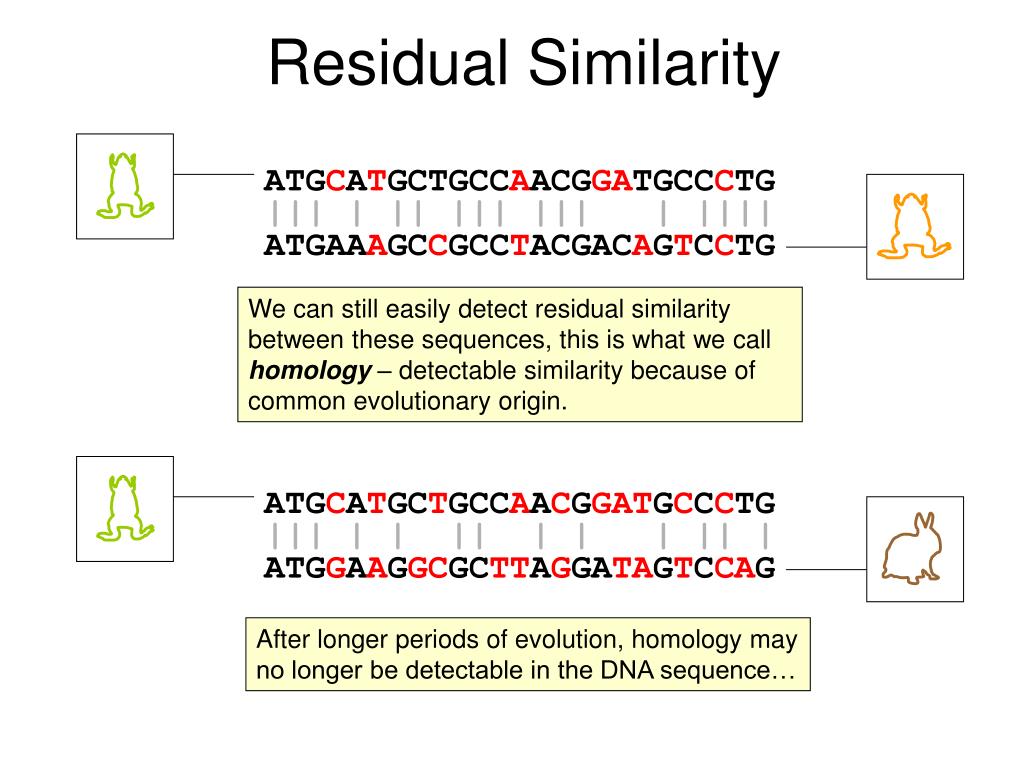
What Is Sequence Identity In Bioinformatics . Fasta, see below) and clustered by similarity scores to produce a guide tree. This module aims to introduce students to the concepts of sequence comparison and identity. Sequence identity is the ratio of the number of identical amino acids between the 2 aligned sequences over the aligned length, expressed as a percentage. The sequences are first compared using a fast method (e.g. Sequence identity is the amount of characters which match exactly between two different sequences. Hereby, gaps are not counted. Sequence identity is the amount of characters which match exactly between two different sequences. Sequence alignment arranges two or more nucleotide or amino acid sequences to identify regions of similarity between the sequences. Comparing dna and protein sequences is a core. Sequence alignment is the process of arranging the characters of a pair of sequences such that the number of matched characters is maximized. Having got the alignment by. We can describe the alignment between.
from www.slideserve.com
Sequence identity is the amount of characters which match exactly between two different sequences. Fasta, see below) and clustered by similarity scores to produce a guide tree. Hereby, gaps are not counted. We can describe the alignment between. Sequence identity is the ratio of the number of identical amino acids between the 2 aligned sequences over the aligned length, expressed as a percentage. The sequences are first compared using a fast method (e.g. Having got the alignment by. Comparing dna and protein sequences is a core. This module aims to introduce students to the concepts of sequence comparison and identity. Sequence identity is the amount of characters which match exactly between two different sequences.
PPT Bioinformatics 1 Sequences and Similarity Searches
What Is Sequence Identity In Bioinformatics Sequence identity is the amount of characters which match exactly between two different sequences. Sequence identity is the amount of characters which match exactly between two different sequences. This module aims to introduce students to the concepts of sequence comparison and identity. The sequences are first compared using a fast method (e.g. Sequence alignment arranges two or more nucleotide or amino acid sequences to identify regions of similarity between the sequences. Sequence alignment is the process of arranging the characters of a pair of sequences such that the number of matched characters is maximized. Sequence identity is the amount of characters which match exactly between two different sequences. Sequence identity is the ratio of the number of identical amino acids between the 2 aligned sequences over the aligned length, expressed as a percentage. We can describe the alignment between. Fasta, see below) and clustered by similarity scores to produce a guide tree. Having got the alignment by. Hereby, gaps are not counted. Comparing dna and protein sequences is a core.
From www.researchgate.net
A. Schematic of the bioinformatics analysis pipeline of data from next What Is Sequence Identity In Bioinformatics This module aims to introduce students to the concepts of sequence comparison and identity. Hereby, gaps are not counted. Sequence identity is the amount of characters which match exactly between two different sequences. The sequences are first compared using a fast method (e.g. We can describe the alignment between. Comparing dna and protein sequences is a core. Sequence alignment arranges. What Is Sequence Identity In Bioinformatics.
From www.slideserve.com
PPT Introduction to Bioinformatics PowerPoint Presentation, free What Is Sequence Identity In Bioinformatics Sequence identity is the amount of characters which match exactly between two different sequences. This module aims to introduce students to the concepts of sequence comparison and identity. Sequence identity is the ratio of the number of identical amino acids between the 2 aligned sequences over the aligned length, expressed as a percentage. Sequence alignment arranges two or more nucleotide. What Is Sequence Identity In Bioinformatics.
From www.youtube.com
What is sequencing depth? Bioinformatics 101 YouTube What Is Sequence Identity In Bioinformatics Sequence identity is the amount of characters which match exactly between two different sequences. Sequence alignment arranges two or more nucleotide or amino acid sequences to identify regions of similarity between the sequences. Comparing dna and protein sequences is a core. This module aims to introduce students to the concepts of sequence comparison and identity. Sequence alignment is the process. What Is Sequence Identity In Bioinformatics.
From www.slideserve.com
PPT CAP5510 Bioinformatics Substitution Patterns PowerPoint What Is Sequence Identity In Bioinformatics Sequence identity is the ratio of the number of identical amino acids between the 2 aligned sequences over the aligned length, expressed as a percentage. Sequence alignment is the process of arranging the characters of a pair of sequences such that the number of matched characters is maximized. Having got the alignment by. Sequence identity is the amount of characters. What Is Sequence Identity In Bioinformatics.
From www.creative-biostructure.com
Protein Sequence Analysis for Coronavirus Research Creative What Is Sequence Identity In Bioinformatics Hereby, gaps are not counted. Having got the alignment by. The sequences are first compared using a fast method (e.g. Comparing dna and protein sequences is a core. Fasta, see below) and clustered by similarity scores to produce a guide tree. This module aims to introduce students to the concepts of sequence comparison and identity. Sequence alignment is the process. What Is Sequence Identity In Bioinformatics.
From www.youtube.com
BIOINFORMATICS_ SEQUENCE ALIGNMENT YouTube What Is Sequence Identity In Bioinformatics Having got the alignment by. Sequence identity is the amount of characters which match exactly between two different sequences. We can describe the alignment between. Sequence identity is the ratio of the number of identical amino acids between the 2 aligned sequences over the aligned length, expressed as a percentage. The sequences are first compared using a fast method (e.g.. What Is Sequence Identity In Bioinformatics.
From www.profacgen.com
Homology Modeling Profacgen What Is Sequence Identity In Bioinformatics Fasta, see below) and clustered by similarity scores to produce a guide tree. The sequences are first compared using a fast method (e.g. We can describe the alignment between. Sequence alignment is the process of arranging the characters of a pair of sequences such that the number of matched characters is maximized. Having got the alignment by. Hereby, gaps are. What Is Sequence Identity In Bioinformatics.
From www.vedantu.com
What is a database in bioinformatics? Mention their two fundamental types. What Is Sequence Identity In Bioinformatics We can describe the alignment between. This module aims to introduce students to the concepts of sequence comparison and identity. Having got the alignment by. The sequences are first compared using a fast method (e.g. Fasta, see below) and clustered by similarity scores to produce a guide tree. Sequence identity is the amount of characters which match exactly between two. What Is Sequence Identity In Bioinformatics.
From www.researchgate.net
Bioinformatics and computational sequence alignment, variant calling What Is Sequence Identity In Bioinformatics Having got the alignment by. We can describe the alignment between. The sequences are first compared using a fast method (e.g. Sequence identity is the ratio of the number of identical amino acids between the 2 aligned sequences over the aligned length, expressed as a percentage. Sequence alignment arranges two or more nucleotide or amino acid sequences to identify regions. What Is Sequence Identity In Bioinformatics.
From www.linstitute.net
IB DP Biology HL复习笔记7.3.4 Bioinformatics翰林国际教育 What Is Sequence Identity In Bioinformatics Sequence identity is the amount of characters which match exactly between two different sequences. Comparing dna and protein sequences is a core. Having got the alignment by. Sequence identity is the ratio of the number of identical amino acids between the 2 aligned sequences over the aligned length, expressed as a percentage. Sequence identity is the amount of characters which. What Is Sequence Identity In Bioinformatics.
From www.researchgate.net
Bioinformatics workflow for handling DNA sequencing data alignment What Is Sequence Identity In Bioinformatics Sequence alignment arranges two or more nucleotide or amino acid sequences to identify regions of similarity between the sequences. Hereby, gaps are not counted. This module aims to introduce students to the concepts of sequence comparison and identity. Comparing dna and protein sequences is a core. Sequence identity is the amount of characters which match exactly between two different sequences.. What Is Sequence Identity In Bioinformatics.
From www.slideserve.com
PPT Bioinformatics Basics PowerPoint Presentation, free download ID What Is Sequence Identity In Bioinformatics Having got the alignment by. Sequence identity is the amount of characters which match exactly between two different sequences. Comparing dna and protein sequences is a core. Sequence identity is the ratio of the number of identical amino acids between the 2 aligned sequences over the aligned length, expressed as a percentage. We can describe the alignment between. The sequences. What Is Sequence Identity In Bioinformatics.
From www.pnnl.gov
Bioinformatics PNNL What Is Sequence Identity In Bioinformatics Sequence identity is the ratio of the number of identical amino acids between the 2 aligned sequences over the aligned length, expressed as a percentage. Having got the alignment by. Comparing dna and protein sequences is a core. This module aims to introduce students to the concepts of sequence comparison and identity. The sequences are first compared using a fast. What Is Sequence Identity In Bioinformatics.
From www.slideserve.com
PPT CSCE555 Bioinformatics PowerPoint Presentation, free download What Is Sequence Identity In Bioinformatics Having got the alignment by. Sequence alignment arranges two or more nucleotide or amino acid sequences to identify regions of similarity between the sequences. Sequence identity is the amount of characters which match exactly between two different sequences. Sequence identity is the ratio of the number of identical amino acids between the 2 aligned sequences over the aligned length, expressed. What Is Sequence Identity In Bioinformatics.
From www.uib.no
Bioinformatics Center for Data Science UiB What Is Sequence Identity In Bioinformatics Sequence alignment arranges two or more nucleotide or amino acid sequences to identify regions of similarity between the sequences. Sequence identity is the amount of characters which match exactly between two different sequences. Fasta, see below) and clustered by similarity scores to produce a guide tree. Comparing dna and protein sequences is a core. We can describe the alignment between.. What Is Sequence Identity In Bioinformatics.
From www.slideserve.com
PPT BCB 444/544 PowerPoint Presentation, free download ID4483547 What Is Sequence Identity In Bioinformatics Fasta, see below) and clustered by similarity scores to produce a guide tree. Sequence identity is the amount of characters which match exactly between two different sequences. Sequence identity is the ratio of the number of identical amino acids between the 2 aligned sequences over the aligned length, expressed as a percentage. Having got the alignment by. The sequences are. What Is Sequence Identity In Bioinformatics.
From www.slideserve.com
PPT Bioinformatics PowerPoint Presentation, free download ID5888868 What Is Sequence Identity In Bioinformatics The sequences are first compared using a fast method (e.g. Fasta, see below) and clustered by similarity scores to produce a guide tree. Sequence alignment is the process of arranging the characters of a pair of sequences such that the number of matched characters is maximized. Sequence identity is the ratio of the number of identical amino acids between the. What Is Sequence Identity In Bioinformatics.
From www.slideserve.com
PPT Bioinformatics Sequence Analysis I PowerPoint Presentation, free What Is Sequence Identity In Bioinformatics Sequence alignment arranges two or more nucleotide or amino acid sequences to identify regions of similarity between the sequences. Comparing dna and protein sequences is a core. This module aims to introduce students to the concepts of sequence comparison and identity. Fasta, see below) and clustered by similarity scores to produce a guide tree. The sequences are first compared using. What Is Sequence Identity In Bioinformatics.
From www.slideserve.com
PPT School of Computer Engineering Master of Science ( Bioinformatics What Is Sequence Identity In Bioinformatics Sequence alignment arranges two or more nucleotide or amino acid sequences to identify regions of similarity between the sequences. We can describe the alignment between. Sequence identity is the amount of characters which match exactly between two different sequences. Sequence alignment is the process of arranging the characters of a pair of sequences such that the number of matched characters. What Is Sequence Identity In Bioinformatics.
From www.biologyexams4u.com
Importance of Sequence Alignment in Bioinformatics What Is Sequence Identity In Bioinformatics Sequence alignment arranges two or more nucleotide or amino acid sequences to identify regions of similarity between the sequences. Hereby, gaps are not counted. The sequences are first compared using a fast method (e.g. This module aims to introduce students to the concepts of sequence comparison and identity. We can describe the alignment between. Comparing dna and protein sequences is. What Is Sequence Identity In Bioinformatics.
From www.slideserve.com
PPT Bioinformatics PowerPoint Presentation, free download ID5888868 What Is Sequence Identity In Bioinformatics Sequence alignment is the process of arranging the characters of a pair of sequences such that the number of matched characters is maximized. Having got the alignment by. Fasta, see below) and clustered by similarity scores to produce a guide tree. This module aims to introduce students to the concepts of sequence comparison and identity. Comparing dna and protein sequences. What Is Sequence Identity In Bioinformatics.
From www.linkedin.com
Sequence Alignment in Bioinformatics What Is Sequence Identity In Bioinformatics The sequences are first compared using a fast method (e.g. Sequence identity is the amount of characters which match exactly between two different sequences. Sequence alignment arranges two or more nucleotide or amino acid sequences to identify regions of similarity between the sequences. Fasta, see below) and clustered by similarity scores to produce a guide tree. Hereby, gaps are not. What Is Sequence Identity In Bioinformatics.
From www.youtube.com
What is the Difference between Primary and Secondary Database in What Is Sequence Identity In Bioinformatics The sequences are first compared using a fast method (e.g. Sequence identity is the ratio of the number of identical amino acids between the 2 aligned sequences over the aligned length, expressed as a percentage. Sequence alignment arranges two or more nucleotide or amino acid sequences to identify regions of similarity between the sequences. Sequence identity is the amount of. What Is Sequence Identity In Bioinformatics.
From www.youtube.com
Sequence Homology, Identity Basic Concepts Explained. YouTube What Is Sequence Identity In Bioinformatics Sequence alignment is the process of arranging the characters of a pair of sequences such that the number of matched characters is maximized. This module aims to introduce students to the concepts of sequence comparison and identity. Sequence identity is the amount of characters which match exactly between two different sequences. The sequences are first compared using a fast method. What Is Sequence Identity In Bioinformatics.
From www.youtube.com
Bioinformatics Sequences Alignment YouTube What Is Sequence Identity In Bioinformatics This module aims to introduce students to the concepts of sequence comparison and identity. Comparing dna and protein sequences is a core. Sequence alignment is the process of arranging the characters of a pair of sequences such that the number of matched characters is maximized. Sequence identity is the amount of characters which match exactly between two different sequences. Sequence. What Is Sequence Identity In Bioinformatics.
From www.slideserve.com
PPT Bioinformatics 1 Sequences and Similarity Searches What Is Sequence Identity In Bioinformatics Sequence identity is the ratio of the number of identical amino acids between the 2 aligned sequences over the aligned length, expressed as a percentage. We can describe the alignment between. Fasta, see below) and clustered by similarity scores to produce a guide tree. Sequence alignment is the process of arranging the characters of a pair of sequences such that. What Is Sequence Identity In Bioinformatics.
From www.researchgate.net
Scheme of the bioinformatics analysis pipeline. Download Scientific What Is Sequence Identity In Bioinformatics Sequence identity is the amount of characters which match exactly between two different sequences. The sequences are first compared using a fast method (e.g. Sequence alignment arranges two or more nucleotide or amino acid sequences to identify regions of similarity between the sequences. Having got the alignment by. We can describe the alignment between. This module aims to introduce students. What Is Sequence Identity In Bioinformatics.
From www.slideserve.com
PPT Sequence Matching and alignment algorithms in the field of What Is Sequence Identity In Bioinformatics Hereby, gaps are not counted. Comparing dna and protein sequences is a core. We can describe the alignment between. Sequence alignment is the process of arranging the characters of a pair of sequences such that the number of matched characters is maximized. The sequences are first compared using a fast method (e.g. This module aims to introduce students to the. What Is Sequence Identity In Bioinformatics.
From www.genomicseducation.hee.nhs.uk
What is bioinformatics? Genomics Education Programme What Is Sequence Identity In Bioinformatics Sequence identity is the amount of characters which match exactly between two different sequences. Sequence identity is the amount of characters which match exactly between two different sequences. Sequence alignment is the process of arranging the characters of a pair of sequences such that the number of matched characters is maximized. Sequence identity is the ratio of the number of. What Is Sequence Identity In Bioinformatics.
From www.slideserve.com
PPT Bioinformatics and sequence analysis PowerPoint Presentation What Is Sequence Identity In Bioinformatics This module aims to introduce students to the concepts of sequence comparison and identity. Having got the alignment by. Comparing dna and protein sequences is a core. Fasta, see below) and clustered by similarity scores to produce a guide tree. The sequences are first compared using a fast method (e.g. Hereby, gaps are not counted. Sequence alignment arranges two or. What Is Sequence Identity In Bioinformatics.
From www.slideserve.com
PPT Development of Bioinformatics and its application on What Is Sequence Identity In Bioinformatics Sequence identity is the amount of characters which match exactly between two different sequences. Hereby, gaps are not counted. Sequence identity is the amount of characters which match exactly between two different sequences. Fasta, see below) and clustered by similarity scores to produce a guide tree. This module aims to introduce students to the concepts of sequence comparison and identity.. What Is Sequence Identity In Bioinformatics.
From www.slideserve.com
PPT Introduction to Bioinformatics PowerPoint Presentation, free What Is Sequence Identity In Bioinformatics We can describe the alignment between. Comparing dna and protein sequences is a core. The sequences are first compared using a fast method (e.g. Hereby, gaps are not counted. Sequence identity is the ratio of the number of identical amino acids between the 2 aligned sequences over the aligned length, expressed as a percentage. Sequence alignment is the process of. What Is Sequence Identity In Bioinformatics.
From www.differencebetween.com
Difference Between Similarity and Identity in Sequence Alignment What Is Sequence Identity In Bioinformatics This module aims to introduce students to the concepts of sequence comparison and identity. Comparing dna and protein sequences is a core. Sequence alignment arranges two or more nucleotide or amino acid sequences to identify regions of similarity between the sequences. Sequence identity is the amount of characters which match exactly between two different sequences. We can describe the alignment. What Is Sequence Identity In Bioinformatics.
From www.slideserve.com
PPT Sequence similarity Analysis PowerPoint Presentation, free What Is Sequence Identity In Bioinformatics Sequence identity is the amount of characters which match exactly between two different sequences. We can describe the alignment between. Having got the alignment by. This module aims to introduce students to the concepts of sequence comparison and identity. Fasta, see below) and clustered by similarity scores to produce a guide tree. Sequence alignment is the process of arranging the. What Is Sequence Identity In Bioinformatics.
From bioinformatics.univ-saida.dz
Sequence Alignment in Bioinformatics What Is Sequence Identity In Bioinformatics This module aims to introduce students to the concepts of sequence comparison and identity. Sequence identity is the ratio of the number of identical amino acids between the 2 aligned sequences over the aligned length, expressed as a percentage. Sequence identity is the amount of characters which match exactly between two different sequences. Sequence alignment is the process of arranging. What Is Sequence Identity In Bioinformatics.