Mass Spectrometry Database Search Engines . Comet is an open source tandem mass spectrometry (ms/ms) sequence database search tool released under the apache 2.0 license. Mascot server is a powerful search engine for identifying proteins and peptides from primary sequence databases using mass spectrometry data. Sequence database search engines are. Searching uninterpreted tandem mass spectra of peptides against sequence databases is the most common method used to identify peptides and. (i) spectra of varying fragmentation methods;
from www.semanticscholar.org
Sequence database search engines are. Searching uninterpreted tandem mass spectra of peptides against sequence databases is the most common method used to identify peptides and. Comet is an open source tandem mass spectrometry (ms/ms) sequence database search tool released under the apache 2.0 license. Mascot server is a powerful search engine for identifying proteins and peptides from primary sequence databases using mass spectrometry data. (i) spectra of varying fragmentation methods;
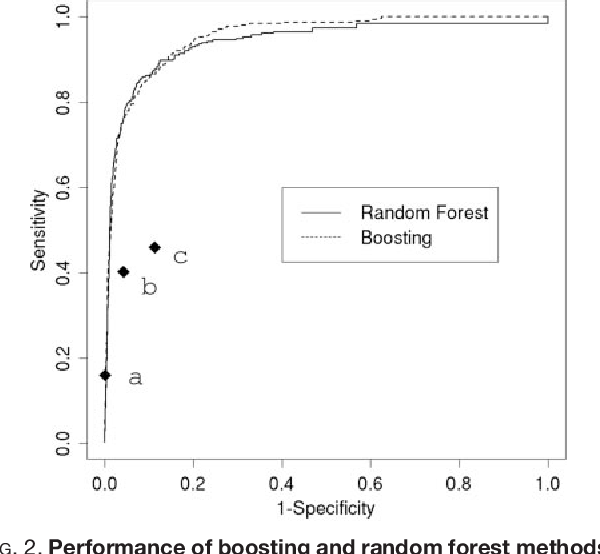
Figure 2 from Improved Classification of Mass Spectrometry Database
Mass Spectrometry Database Search Engines Sequence database search engines are. Searching uninterpreted tandem mass spectra of peptides against sequence databases is the most common method used to identify peptides and. (i) spectra of varying fragmentation methods; Comet is an open source tandem mass spectrometry (ms/ms) sequence database search tool released under the apache 2.0 license. Mascot server is a powerful search engine for identifying proteins and peptides from primary sequence databases using mass spectrometry data. Sequence database search engines are.
From www.npsciences.com
Nordic Preclinical Sciences Proteomics Analysis Mass Spectroscopy Mass Spectrometry Database Search Engines Sequence database search engines are. (i) spectra of varying fragmentation methods; Comet is an open source tandem mass spectrometry (ms/ms) sequence database search tool released under the apache 2.0 license. Mascot server is a powerful search engine for identifying proteins and peptides from primary sequence databases using mass spectrometry data. Searching uninterpreted tandem mass spectra of peptides against sequence databases. Mass Spectrometry Database Search Engines.
From ai.fiu.edu
SpeCollate Deep CrossModal Similarity Network for Peptide Database Mass Spectrometry Database Search Engines Searching uninterpreted tandem mass spectra of peptides against sequence databases is the most common method used to identify peptides and. Sequence database search engines are. Mascot server is a powerful search engine for identifying proteins and peptides from primary sequence databases using mass spectrometry data. (i) spectra of varying fragmentation methods; Comet is an open source tandem mass spectrometry (ms/ms). Mass Spectrometry Database Search Engines.
From www.semanticscholar.org
Table 2 from Anatomy and evolution of database search enginesa central Mass Spectrometry Database Search Engines (i) spectra of varying fragmentation methods; Searching uninterpreted tandem mass spectra of peptides against sequence databases is the most common method used to identify peptides and. Sequence database search engines are. Comet is an open source tandem mass spectrometry (ms/ms) sequence database search tool released under the apache 2.0 license. Mascot server is a powerful search engine for identifying proteins. Mass Spectrometry Database Search Engines.
From www.bronkhorst.com
Mass Spectrometry and Mass Flow Control; A closer ion them Bronkhorst Mass Spectrometry Database Search Engines (i) spectra of varying fragmentation methods; Mascot server is a powerful search engine for identifying proteins and peptides from primary sequence databases using mass spectrometry data. Searching uninterpreted tandem mass spectra of peptides against sequence databases is the most common method used to identify peptides and. Comet is an open source tandem mass spectrometry (ms/ms) sequence database search tool released. Mass Spectrometry Database Search Engines.
From www.nist.gov
A screenshot of the NIST Mass Spectral Library user interface Mass Spectrometry Database Search Engines Searching uninterpreted tandem mass spectra of peptides against sequence databases is the most common method used to identify peptides and. (i) spectra of varying fragmentation methods; Mascot server is a powerful search engine for identifying proteins and peptides from primary sequence databases using mass spectrometry data. Comet is an open source tandem mass spectrometry (ms/ms) sequence database search tool released. Mass Spectrometry Database Search Engines.
From www.umassmed.edu
Thermo Scientific Q Exactive Plus Orbitrap Mass Spectrometer Mass Spectrometry Database Search Engines Mascot server is a powerful search engine for identifying proteins and peptides from primary sequence databases using mass spectrometry data. (i) spectra of varying fragmentation methods; Searching uninterpreted tandem mass spectra of peptides against sequence databases is the most common method used to identify peptides and. Sequence database search engines are. Comet is an open source tandem mass spectrometry (ms/ms). Mass Spectrometry Database Search Engines.
From github.com
GitHub wasimsandhu/SearchMS Search engine for mass spectrometry Mass Spectrometry Database Search Engines Sequence database search engines are. Searching uninterpreted tandem mass spectra of peptides against sequence databases is the most common method used to identify peptides and. Comet is an open source tandem mass spectrometry (ms/ms) sequence database search tool released under the apache 2.0 license. Mascot server is a powerful search engine for identifying proteins and peptides from primary sequence databases. Mass Spectrometry Database Search Engines.
From sciencesolutions.wiley.com
Mass Spectral Databases Wiley Science Solutions Mass Spectrometry Database Search Engines (i) spectra of varying fragmentation methods; Mascot server is a powerful search engine for identifying proteins and peptides from primary sequence databases using mass spectrometry data. Searching uninterpreted tandem mass spectra of peptides against sequence databases is the most common method used to identify peptides and. Comet is an open source tandem mass spectrometry (ms/ms) sequence database search tool released. Mass Spectrometry Database Search Engines.
From www.sisweb.com
NIST 20 Mass Spectral Library, NIST 2020/2017 Database, Agilent Format Mass Spectrometry Database Search Engines Comet is an open source tandem mass spectrometry (ms/ms) sequence database search tool released under the apache 2.0 license. Sequence database search engines are. Searching uninterpreted tandem mass spectra of peptides against sequence databases is the most common method used to identify peptides and. (i) spectra of varying fragmentation methods; Mascot server is a powerful search engine for identifying proteins. Mass Spectrometry Database Search Engines.
From ms.epfl.ch
Mass Spectrometry database online The ISIC EPFL mstoolbox Mass Spectrometry Database Search Engines Comet is an open source tandem mass spectrometry (ms/ms) sequence database search tool released under the apache 2.0 license. Mascot server is a powerful search engine for identifying proteins and peptides from primary sequence databases using mass spectrometry data. Searching uninterpreted tandem mass spectra of peptides against sequence databases is the most common method used to identify peptides and. Sequence. Mass Spectrometry Database Search Engines.
From www.researchgate.net
Workflow for identifying gene fusion and splicing events from MS/MS Mass Spectrometry Database Search Engines (i) spectra of varying fragmentation methods; Mascot server is a powerful search engine for identifying proteins and peptides from primary sequence databases using mass spectrometry data. Sequence database search engines are. Searching uninterpreted tandem mass spectra of peptides against sequence databases is the most common method used to identify peptides and. Comet is an open source tandem mass spectrometry (ms/ms). Mass Spectrometry Database Search Engines.
From blog.sepscience.com
FREE Master Class on Effective Use of Mass Spectral Databases Mass Spectrometry Database Search Engines Comet is an open source tandem mass spectrometry (ms/ms) sequence database search tool released under the apache 2.0 license. Searching uninterpreted tandem mass spectra of peptides against sequence databases is the most common method used to identify peptides and. Sequence database search engines are. Mascot server is a powerful search engine for identifying proteins and peptides from primary sequence databases. Mass Spectrometry Database Search Engines.
From pages.nist.gov
User Guide for the NIST Database Infrastructure for Mass Spectrometry Mass Spectrometry Database Search Engines Comet is an open source tandem mass spectrometry (ms/ms) sequence database search tool released under the apache 2.0 license. (i) spectra of varying fragmentation methods; Mascot server is a powerful search engine for identifying proteins and peptides from primary sequence databases using mass spectrometry data. Sequence database search engines are. Searching uninterpreted tandem mass spectra of peptides against sequence databases. Mass Spectrometry Database Search Engines.
From www.spectroscopyeurope.com
New mass spectrometry method for characterisation of the most Mass Spectrometry Database Search Engines Comet is an open source tandem mass spectrometry (ms/ms) sequence database search tool released under the apache 2.0 license. Mascot server is a powerful search engine for identifying proteins and peptides from primary sequence databases using mass spectrometry data. Searching uninterpreted tandem mass spectra of peptides against sequence databases is the most common method used to identify peptides and. Sequence. Mass Spectrometry Database Search Engines.
From www.semanticscholar.org
Figure 2 from Improved Classification of Mass Spectrometry Database Mass Spectrometry Database Search Engines Comet is an open source tandem mass spectrometry (ms/ms) sequence database search tool released under the apache 2.0 license. Mascot server is a powerful search engine for identifying proteins and peptides from primary sequence databases using mass spectrometry data. Sequence database search engines are. Searching uninterpreted tandem mass spectra of peptides against sequence databases is the most common method used. Mass Spectrometry Database Search Engines.
From www.researchgate.net
Mass Spectra Database for Products of Glucose Download Mass Spectrometry Database Search Engines (i) spectra of varying fragmentation methods; Mascot server is a powerful search engine for identifying proteins and peptides from primary sequence databases using mass spectrometry data. Sequence database search engines are. Searching uninterpreted tandem mass spectra of peptides against sequence databases is the most common method used to identify peptides and. Comet is an open source tandem mass spectrometry (ms/ms). Mass Spectrometry Database Search Engines.
From pubs.acs.org
An Introduction to Mass SpectrometryBased Proteomics Journal of Mass Spectrometry Database Search Engines Comet is an open source tandem mass spectrometry (ms/ms) sequence database search tool released under the apache 2.0 license. Mascot server is a powerful search engine for identifying proteins and peptides from primary sequence databases using mass spectrometry data. (i) spectra of varying fragmentation methods; Sequence database search engines are. Searching uninterpreted tandem mass spectra of peptides against sequence databases. Mass Spectrometry Database Search Engines.
From journal.frontiersin.org
Frontiers ProMEX a mass spectral reference database for plant Mass Spectrometry Database Search Engines (i) spectra of varying fragmentation methods; Sequence database search engines are. Mascot server is a powerful search engine for identifying proteins and peptides from primary sequence databases using mass spectrometry data. Searching uninterpreted tandem mass spectra of peptides against sequence databases is the most common method used to identify peptides and. Comet is an open source tandem mass spectrometry (ms/ms). Mass Spectrometry Database Search Engines.
From www.researchgate.net
Anatomy and evolution of database search engines—a central component of Mass Spectrometry Database Search Engines (i) spectra of varying fragmentation methods; Mascot server is a powerful search engine for identifying proteins and peptides from primary sequence databases using mass spectrometry data. Searching uninterpreted tandem mass spectra of peptides against sequence databases is the most common method used to identify peptides and. Sequence database search engines are. Comet is an open source tandem mass spectrometry (ms/ms). Mass Spectrometry Database Search Engines.
From chemrxiv.org
Proteomic Database Search Engine for TwoDimensional Partial Covariance Mass Spectrometry Database Search Engines Sequence database search engines are. Searching uninterpreted tandem mass spectra of peptides against sequence databases is the most common method used to identify peptides and. (i) spectra of varying fragmentation methods; Mascot server is a powerful search engine for identifying proteins and peptides from primary sequence databases using mass spectrometry data. Comet is an open source tandem mass spectrometry (ms/ms). Mass Spectrometry Database Search Engines.
From analyticalsciencejournals.onlinelibrary.wiley.com
Anatomy and evolution of database search engines—a central component of Mass Spectrometry Database Search Engines Sequence database search engines are. Searching uninterpreted tandem mass spectra of peptides against sequence databases is the most common method used to identify peptides and. (i) spectra of varying fragmentation methods; Mascot server is a powerful search engine for identifying proteins and peptides from primary sequence databases using mass spectrometry data. Comet is an open source tandem mass spectrometry (ms/ms). Mass Spectrometry Database Search Engines.
From www.researchgate.net
Data acquisition and database search. For a given experimental MS/MS Mass Spectrometry Database Search Engines Searching uninterpreted tandem mass spectra of peptides against sequence databases is the most common method used to identify peptides and. Sequence database search engines are. (i) spectra of varying fragmentation methods; Mascot server is a powerful search engine for identifying proteins and peptides from primary sequence databases using mass spectrometry data. Comet is an open source tandem mass spectrometry (ms/ms). Mass Spectrometry Database Search Engines.
From www.slideserve.com
PPT PepHMM A Hidden Markov Model Based Scoring Function for Mass Mass Spectrometry Database Search Engines Mascot server is a powerful search engine for identifying proteins and peptides from primary sequence databases using mass spectrometry data. Sequence database search engines are. (i) spectra of varying fragmentation methods; Searching uninterpreted tandem mass spectra of peptides against sequence databases is the most common method used to identify peptides and. Comet is an open source tandem mass spectrometry (ms/ms). Mass Spectrometry Database Search Engines.
From www.researchgate.net
Data acquisition and database search. For a given experimental MS/MS Mass Spectrometry Database Search Engines Mascot server is a powerful search engine for identifying proteins and peptides from primary sequence databases using mass spectrometry data. Comet is an open source tandem mass spectrometry (ms/ms) sequence database search tool released under the apache 2.0 license. Sequence database search engines are. Searching uninterpreted tandem mass spectra of peptides against sequence databases is the most common method used. Mass Spectrometry Database Search Engines.
From www.semanticscholar.org
Figure 1 from Anatomy and evolution of database search enginesa Mass Spectrometry Database Search Engines (i) spectra of varying fragmentation methods; Searching uninterpreted tandem mass spectra of peptides against sequence databases is the most common method used to identify peptides and. Sequence database search engines are. Comet is an open source tandem mass spectrometry (ms/ms) sequence database search tool released under the apache 2.0 license. Mascot server is a powerful search engine for identifying proteins. Mass Spectrometry Database Search Engines.
From www.semanticscholar.org
Figure 3 from Anatomy and evolution of database search enginesa Mass Spectrometry Database Search Engines Comet is an open source tandem mass spectrometry (ms/ms) sequence database search tool released under the apache 2.0 license. Sequence database search engines are. Mascot server is a powerful search engine for identifying proteins and peptides from primary sequence databases using mass spectrometry data. (i) spectra of varying fragmentation methods; Searching uninterpreted tandem mass spectra of peptides against sequence databases. Mass Spectrometry Database Search Engines.
From www.semanticscholar.org
Figure 4 from Anatomy and evolution of database search enginesa Mass Spectrometry Database Search Engines Searching uninterpreted tandem mass spectra of peptides against sequence databases is the most common method used to identify peptides and. Mascot server is a powerful search engine for identifying proteins and peptides from primary sequence databases using mass spectrometry data. Sequence database search engines are. (i) spectra of varying fragmentation methods; Comet is an open source tandem mass spectrometry (ms/ms). Mass Spectrometry Database Search Engines.
From mavink.com
Mass Spectrometry Chart Mass Spectrometry Database Search Engines Searching uninterpreted tandem mass spectra of peptides against sequence databases is the most common method used to identify peptides and. Mascot server is a powerful search engine for identifying proteins and peptides from primary sequence databases using mass spectrometry data. (i) spectra of varying fragmentation methods; Comet is an open source tandem mass spectrometry (ms/ms) sequence database search tool released. Mass Spectrometry Database Search Engines.
From www.mdpi.com
IJMS Free FullText Bioinformatics Methods for Mass Spectrometry Mass Spectrometry Database Search Engines Comet is an open source tandem mass spectrometry (ms/ms) sequence database search tool released under the apache 2.0 license. Searching uninterpreted tandem mass spectra of peptides against sequence databases is the most common method used to identify peptides and. (i) spectra of varying fragmentation methods; Mascot server is a powerful search engine for identifying proteins and peptides from primary sequence. Mass Spectrometry Database Search Engines.
From www.researchgate.net
Summary of Important Mass Spectral Databases Download Table Mass Spectrometry Database Search Engines Sequence database search engines are. Searching uninterpreted tandem mass spectra of peptides against sequence databases is the most common method used to identify peptides and. Mascot server is a powerful search engine for identifying proteins and peptides from primary sequence databases using mass spectrometry data. (i) spectra of varying fragmentation methods; Comet is an open source tandem mass spectrometry (ms/ms). Mass Spectrometry Database Search Engines.
From www.semanticscholar.org
Anatomy and evolution of database search enginesa central component of Mass Spectrometry Database Search Engines Comet is an open source tandem mass spectrometry (ms/ms) sequence database search tool released under the apache 2.0 license. Mascot server is a powerful search engine for identifying proteins and peptides from primary sequence databases using mass spectrometry data. Sequence database search engines are. Searching uninterpreted tandem mass spectra of peptides against sequence databases is the most common method used. Mass Spectrometry Database Search Engines.
From pages.nist.gov
User Guide for the NIST Database Infrastructure for Mass Spectrometry Mass Spectrometry Database Search Engines Comet is an open source tandem mass spectrometry (ms/ms) sequence database search tool released under the apache 2.0 license. (i) spectra of varying fragmentation methods; Sequence database search engines are. Searching uninterpreted tandem mass spectra of peptides against sequence databases is the most common method used to identify peptides and. Mascot server is a powerful search engine for identifying proteins. Mass Spectrometry Database Search Engines.
From www.semanticscholar.org
[PDF] Improved Classification of Mass Spectrometry Database Search Mass Spectrometry Database Search Engines Searching uninterpreted tandem mass spectra of peptides against sequence databases is the most common method used to identify peptides and. Comet is an open source tandem mass spectrometry (ms/ms) sequence database search tool released under the apache 2.0 license. Mascot server is a powerful search engine for identifying proteins and peptides from primary sequence databases using mass spectrometry data. Sequence. Mass Spectrometry Database Search Engines.
From ms.epfl.ch
Mass Spectrometry database online The ISIC EPFL mstoolbox Mass Spectrometry Database Search Engines Searching uninterpreted tandem mass spectra of peptides against sequence databases is the most common method used to identify peptides and. Comet is an open source tandem mass spectrometry (ms/ms) sequence database search tool released under the apache 2.0 license. Mascot server is a powerful search engine for identifying proteins and peptides from primary sequence databases using mass spectrometry data. (i). Mass Spectrometry Database Search Engines.
From www.researchgate.net
Identification of sorcin by mass spectrometry analysis and database Mass Spectrometry Database Search Engines Searching uninterpreted tandem mass spectra of peptides against sequence databases is the most common method used to identify peptides and. Comet is an open source tandem mass spectrometry (ms/ms) sequence database search tool released under the apache 2.0 license. Sequence database search engines are. (i) spectra of varying fragmentation methods; Mascot server is a powerful search engine for identifying proteins. Mass Spectrometry Database Search Engines.