Sequencing Bam File . Sam and bam files are text and binary formats for storing alignment data of nucleic acid sequences. Learn what bam files are, how they are created, and how they store information about aligned reads. Bam files are binary, compressed, indexed versions of sam files, a common output. Find out how to load, filter, and annotate. Learn how to sort, index and analyze sam and bam files with bioinformatics. This web page introduces the bam format, how to install and use. Bam is a binary format for storing large nucleotide sequence alignments. Learn what a bam file is, how it is compressed from a sam file, and what information it contains. A bam file is used to represent aligned. Learn how to use igv to visualise and interpret bam files, which store alignment data of sequencing reads to a reference genome.
from hbctraining.github.io
A bam file is used to represent aligned. Learn what a bam file is, how it is compressed from a sam file, and what information it contains. Sam and bam files are text and binary formats for storing alignment data of nucleic acid sequences. Learn how to use igv to visualise and interpret bam files, which store alignment data of sequencing reads to a reference genome. Bam is a binary format for storing large nucleotide sequence alignments. Bam files are binary, compressed, indexed versions of sam files, a common output. Find out how to load, filter, and annotate. Learn what bam files are, how they are created, and how they store information about aligned reads. This web page introduces the bam format, how to install and use. Learn how to sort, index and analyze sam and bam files with bioinformatics.
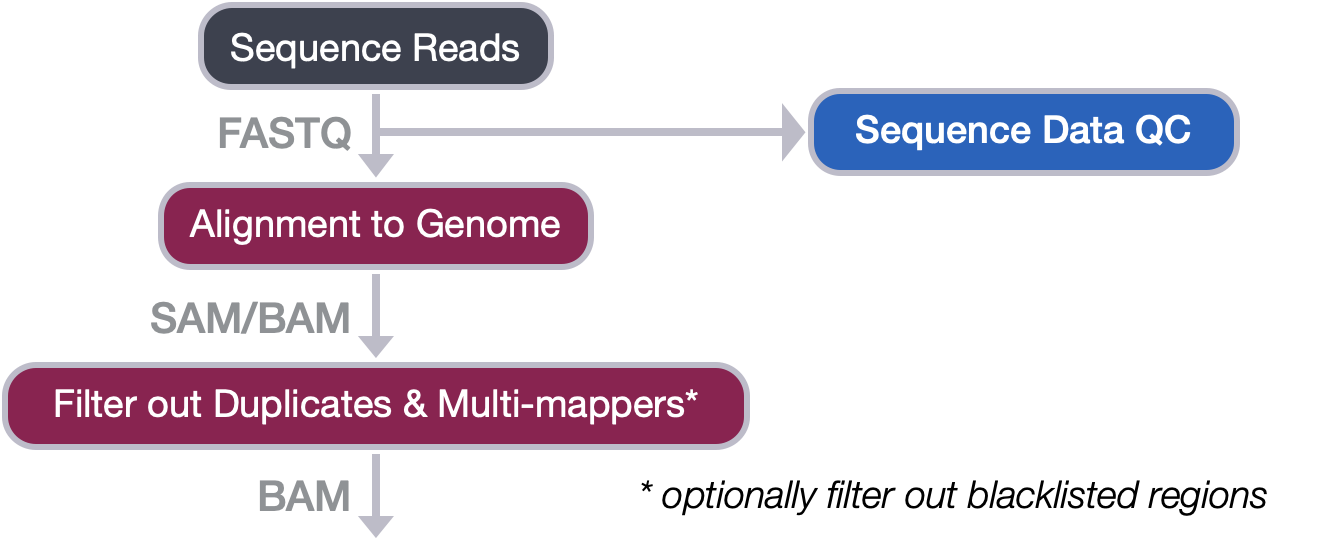
Filtering BAM files Introduction to ChIPSeq using highperformance
Sequencing Bam File A bam file is used to represent aligned. Bam is a binary format for storing large nucleotide sequence alignments. Learn what bam files are, how they are created, and how they store information about aligned reads. Learn how to use igv to visualise and interpret bam files, which store alignment data of sequencing reads to a reference genome. Learn how to sort, index and analyze sam and bam files with bioinformatics. Bam files are binary, compressed, indexed versions of sam files, a common output. Sam and bam files are text and binary formats for storing alignment data of nucleic acid sequences. A bam file is used to represent aligned. Find out how to load, filter, and annotate. This web page introduces the bam format, how to install and use. Learn what a bam file is, how it is compressed from a sam file, and what information it contains.
From blog.liang2.tw
Plot Sequencing Depth with Gviz Sequencing Bam File Find out how to load, filter, and annotate. Learn what bam files are, how they are created, and how they store information about aligned reads. Learn how to sort, index and analyze sam and bam files with bioinformatics. Learn what a bam file is, how it is compressed from a sam file, and what information it contains. Learn how to. Sequencing Bam File.
From www.researchgate.net
Overview of the NeoSplice method. BAM files from tumor and Sequencing Bam File Learn how to sort, index and analyze sam and bam files with bioinformatics. A bam file is used to represent aligned. Learn how to use igv to visualise and interpret bam files, which store alignment data of sequencing reads to a reference genome. Bam files are binary, compressed, indexed versions of sam files, a common output. Bam is a binary. Sequencing Bam File.
From www.pdfprof.com
cram file to bam file Sequencing Bam File Learn what a bam file is, how it is compressed from a sam file, and what information it contains. A bam file is used to represent aligned. Bam files are binary, compressed, indexed versions of sam files, a common output. Bam is a binary format for storing large nucleotide sequence alignments. Sam and bam files are text and binary formats. Sequencing Bam File.
From hbctraining.github.io
Filtering BAM files Introduction to ChIPSeq using highperformance Sequencing Bam File Learn how to sort, index and analyze sam and bam files with bioinformatics. A bam file is used to represent aligned. Learn what bam files are, how they are created, and how they store information about aligned reads. Bam is a binary format for storing large nucleotide sequence alignments. Learn how to use igv to visualise and interpret bam files,. Sequencing Bam File.
From ovasgpainting.weebly.com
Cigar bam file format description ovasgpainting Sequencing Bam File This web page introduces the bam format, how to install and use. A bam file is used to represent aligned. Sam and bam files are text and binary formats for storing alignment data of nucleic acid sequences. Bam files are binary, compressed, indexed versions of sam files, a common output. Learn how to sort, index and analyze sam and bam. Sequencing Bam File.
From learn.gencore.bio.nyu.edu
NGS Sequencing Technology and File Formats NGS Analysis Sequencing Bam File Sam and bam files are text and binary formats for storing alignment data of nucleic acid sequences. This web page introduces the bam format, how to install and use. Find out how to load, filter, and annotate. Learn what a bam file is, how it is compressed from a sam file, and what information it contains. Bam is a binary. Sequencing Bam File.
From hbctraining.github.io
SAM/BAM file and assessing quality Introduction to RNASeq using high Sequencing Bam File Bam files are binary, compressed, indexed versions of sam files, a common output. Learn what a bam file is, how it is compressed from a sam file, and what information it contains. Learn how to sort, index and analyze sam and bam files with bioinformatics. Find out how to load, filter, and annotate. Bam is a binary format for storing. Sequencing Bam File.
From learngenomics.dev
BAM Files Introduction to Genomics for Engineers Sequencing Bam File Find out how to load, filter, and annotate. A bam file is used to represent aligned. Learn how to use igv to visualise and interpret bam files, which store alignment data of sequencing reads to a reference genome. Bam files are binary, compressed, indexed versions of sam files, a common output. Learn what bam files are, how they are created,. Sequencing Bam File.
From paleogenomics-tor-vergata.readthedocs.io
7. Alignment of reads to a reference genome — Paleogenomics at Tor Sequencing Bam File Bam is a binary format for storing large nucleotide sequence alignments. Find out how to load, filter, and annotate. Sam and bam files are text and binary formats for storing alignment data of nucleic acid sequences. Learn how to sort, index and analyze sam and bam files with bioinformatics. Learn how to use igv to visualise and interpret bam files,. Sequencing Bam File.
From www.researchgate.net
How do I create a consensus fasta sequence from longread sequencing Sequencing Bam File Learn how to sort, index and analyze sam and bam files with bioinformatics. Find out how to load, filter, and annotate. Sam and bam files are text and binary formats for storing alignment data of nucleic acid sequences. Learn how to use igv to visualise and interpret bam files, which store alignment data of sequencing reads to a reference genome.. Sequencing Bam File.
From www.researchgate.net
Workflow of APAScan. Starting with aligned RNAseq and 3'endseq Sequencing Bam File A bam file is used to represent aligned. Learn how to use igv to visualise and interpret bam files, which store alignment data of sequencing reads to a reference genome. This web page introduces the bam format, how to install and use. Learn what bam files are, how they are created, and how they store information about aligned reads. Find. Sequencing Bam File.
From learn.gencore.bio.nyu.edu
Deeptools2 bamCoverage NGS Analysis Sequencing Bam File Learn what a bam file is, how it is compressed from a sam file, and what information it contains. Learn how to use igv to visualise and interpret bam files, which store alignment data of sequencing reads to a reference genome. Bam is a binary format for storing large nucleotide sequence alignments. Sam and bam files are text and binary. Sequencing Bam File.
From hbctraining.github.io
Filtering BAM files Introduction to ChIPSeq using highperformance Sequencing Bam File Learn how to use igv to visualise and interpret bam files, which store alignment data of sequencing reads to a reference genome. Learn how to sort, index and analyze sam and bam files with bioinformatics. This web page introduces the bam format, how to install and use. A bam file is used to represent aligned. Find out how to load,. Sequencing Bam File.
From sepsis-omics.github.io
Viral genome sequencing ABRPITraining Sequencing Bam File A bam file is used to represent aligned. Bam is a binary format for storing large nucleotide sequence alignments. Learn how to use igv to visualise and interpret bam files, which store alignment data of sequencing reads to a reference genome. This web page introduces the bam format, how to install and use. Find out how to load, filter, and. Sequencing Bam File.
From dasegerman.weebly.com
Posorted bam file format 10x scatac dasegerman Sequencing Bam File Learn how to use igv to visualise and interpret bam files, which store alignment data of sequencing reads to a reference genome. Bam is a binary format for storing large nucleotide sequence alignments. Find out how to load, filter, and annotate. Bam files are binary, compressed, indexed versions of sam files, a common output. Sam and bam files are text. Sequencing Bam File.
From nbis-workshop-epigenomics.readthedocs.io
Quality Control for ATACseq — Epigenomics 2024 1 documentation Sequencing Bam File This web page introduces the bam format, how to install and use. Bam files are binary, compressed, indexed versions of sam files, a common output. Find out how to load, filter, and annotate. Learn what a bam file is, how it is compressed from a sam file, and what information it contains. A bam file is used to represent aligned.. Sequencing Bam File.
From www.researchgate.net
Workflow of ScanNeo. Starting with aligned RNASeq BAM file, ScanNeo Sequencing Bam File A bam file is used to represent aligned. Learn how to sort, index and analyze sam and bam files with bioinformatics. Bam files are binary, compressed, indexed versions of sam files, a common output. Learn what bam files are, how they are created, and how they store information about aligned reads. Sam and bam files are text and binary formats. Sequencing Bam File.
From scienceparkstudygroup.github.io
03 From fastq files to alignments Introduction to RNAseq Sequencing Bam File Learn what bam files are, how they are created, and how they store information about aligned reads. Learn how to use igv to visualise and interpret bam files, which store alignment data of sequencing reads to a reference genome. Bam files are binary, compressed, indexed versions of sam files, a common output. Find out how to load, filter, and annotate.. Sequencing Bam File.
From www.xiahepublishing.com
Clinical Application of NGS Tools in the Diagnosis of Collagenopathies Sequencing Bam File Learn how to sort, index and analyze sam and bam files with bioinformatics. Learn how to use igv to visualise and interpret bam files, which store alignment data of sequencing reads to a reference genome. A bam file is used to represent aligned. This web page introduces the bam format, how to install and use. Bam is a binary format. Sequencing Bam File.
From seandavi.github.io
ATACSeq with Bioconductor • Sequencing Bam File Bam is a binary format for storing large nucleotide sequence alignments. A bam file is used to represent aligned. Sam and bam files are text and binary formats for storing alignment data of nucleic acid sequences. Bam files are binary, compressed, indexed versions of sam files, a common output. Learn what bam files are, how they are created, and how. Sequencing Bam File.
From scfbm.biomedcentral.com
Biosamtools Ruby bindings for SAMtools, a library for accessing BAM Sequencing Bam File Sam and bam files are text and binary formats for storing alignment data of nucleic acid sequences. Learn how to use igv to visualise and interpret bam files, which store alignment data of sequencing reads to a reference genome. Bam is a binary format for storing large nucleotide sequence alignments. A bam file is used to represent aligned. Learn how. Sequencing Bam File.
From sites.psu.edu
Good ways to visualize aligned small RNAseq data Axtell Lab at Penn Sequencing Bam File Learn what a bam file is, how it is compressed from a sam file, and what information it contains. This web page introduces the bam format, how to install and use. Learn what bam files are, how they are created, and how they store information about aligned reads. Bam is a binary format for storing large nucleotide sequence alignments. Find. Sequencing Bam File.
From learndarelo.weebly.com
bam file format sequencing data learndarelo Sequencing Bam File Learn what a bam file is, how it is compressed from a sam file, and what information it contains. Learn how to sort, index and analyze sam and bam files with bioinformatics. Sam and bam files are text and binary formats for storing alignment data of nucleic acid sequences. Learn what bam files are, how they are created, and how. Sequencing Bam File.
From texaspsado.weebly.com
bam file format sequencing data texaspsado Sequencing Bam File Learn how to sort, index and analyze sam and bam files with bioinformatics. Sam and bam files are text and binary formats for storing alignment data of nucleic acid sequences. Learn what bam files are, how they are created, and how they store information about aligned reads. A bam file is used to represent aligned. Learn what a bam file. Sequencing Bam File.
From www.youtube.com
Genomic file formats BAM / SAM file format YouTube Sequencing Bam File Sam and bam files are text and binary formats for storing alignment data of nucleic acid sequences. Learn what a bam file is, how it is compressed from a sam file, and what information it contains. Learn how to sort, index and analyze sam and bam files with bioinformatics. Bam files are binary, compressed, indexed versions of sam files, a. Sequencing Bam File.
From ucdavis-bioinformatics-training.github.io
Alignment to Read Counts & Visualization in IGV UC Davis Sequencing Bam File Bam is a binary format for storing large nucleotide sequence alignments. This web page introduces the bam format, how to install and use. Find out how to load, filter, and annotate. Learn what bam files are, how they are created, and how they store information about aligned reads. Sam and bam files are text and binary formats for storing alignment. Sequencing Bam File.
From blog.liang2.tw
Plot Sequencing Depth with Gviz Sequencing Bam File Bam files are binary, compressed, indexed versions of sam files, a common output. Learn how to sort, index and analyze sam and bam files with bioinformatics. Learn what a bam file is, how it is compressed from a sam file, and what information it contains. Learn how to use igv to visualise and interpret bam files, which store alignment data. Sequencing Bam File.
From qlucore.com
Import RNAseq BAM files Qlucore Sequencing Bam File Sam and bam files are text and binary formats for storing alignment data of nucleic acid sequences. Learn how to use igv to visualise and interpret bam files, which store alignment data of sequencing reads to a reference genome. This web page introduces the bam format, how to install and use. Learn what a bam file is, how it is. Sequencing Bam File.
From www.researchgate.net
Workflow of ASQuant. Starting with aligned RNAseq bam files, ASQuant Sequencing Bam File Find out how to load, filter, and annotate. A bam file is used to represent aligned. Sam and bam files are text and binary formats for storing alignment data of nucleic acid sequences. Learn what a bam file is, how it is compressed from a sam file, and what information it contains. This web page introduces the bam format, how. Sequencing Bam File.
From www.biostars.org
Why does TCGA RNAseq pipeline starts from a BAM file? Sequencing Bam File Learn how to use igv to visualise and interpret bam files, which store alignment data of sequencing reads to a reference genome. Bam files are binary, compressed, indexed versions of sam files, a common output. Learn what bam files are, how they are created, and how they store information about aligned reads. Learn what a bam file is, how it. Sequencing Bam File.
From scnbase.org
How do I upload a bam file? SCNBase Sequencing Bam File Bam is a binary format for storing large nucleotide sequence alignments. Learn how to use igv to visualise and interpret bam files, which store alignment data of sequencing reads to a reference genome. Bam files are binary, compressed, indexed versions of sam files, a common output. A bam file is used to represent aligned. Sam and bam files are text. Sequencing Bam File.
From www.researchgate.net
Reanalysis of RNAseq data. a Overview of BAMscale RNAseq mode Sequencing Bam File Learn what a bam file is, how it is compressed from a sam file, and what information it contains. Bam is a binary format for storing large nucleotide sequence alignments. This web page introduces the bam format, how to install and use. Find out how to load, filter, and annotate. A bam file is used to represent aligned. Learn how. Sequencing Bam File.
From www.researchgate.net
Alignment of bam files of the two growth types in IGV. yb144 052144 Sequencing Bam File Bam is a binary format for storing large nucleotide sequence alignments. Bam files are binary, compressed, indexed versions of sam files, a common output. Learn what a bam file is, how it is compressed from a sam file, and what information it contains. Learn how to sort, index and analyze sam and bam files with bioinformatics. This web page introduces. Sequencing Bam File.
From www.biobam.com
BAM File Quality Control BioBam Sequencing Bam File Learn how to use igv to visualise and interpret bam files, which store alignment data of sequencing reads to a reference genome. Learn how to sort, index and analyze sam and bam files with bioinformatics. Learn what a bam file is, how it is compressed from a sam file, and what information it contains. Bam files are binary, compressed, indexed. Sequencing Bam File.
From www.researchgate.net
of the PEAS framework. Features were extracted from ATACseq bam files Sequencing Bam File Learn what bam files are, how they are created, and how they store information about aligned reads. Find out how to load, filter, and annotate. Learn how to use igv to visualise and interpret bam files, which store alignment data of sequencing reads to a reference genome. Learn how to sort, index and analyze sam and bam files with bioinformatics.. Sequencing Bam File.