Trim Galore Biostars . Cutadapt has the parameter setting that asking you to. I'm trying to run trim galore on multiple files (24), but i haven't gotten it to work. Is a wrapper script to automate quality and adapter trimming as well as quality control, with some added functionality to. Trim galore is a wrapper around cutadapt and fastqc to consistently apply adapter and quality trimming to fastq files, with extra functionality for rrbs data. As i understand, trim galore! The code i'm using is: By default, trim galore removes even a single bp match to adapter seq. So some spots may have only one read after. These results are _trimmed.fq files. Which tool is the best to trim the raw data?
from blog.csdn.net
So some spots may have only one read after. The code i'm using is: Is a wrapper script to automate quality and adapter trimming as well as quality control, with some added functionality to. By default, trim galore removes even a single bp match to adapter seq. As i understand, trim galore! Which tool is the best to trim the raw data? Cutadapt has the parameter setting that asking you to. I'm trying to run trim galore on multiple files (24), but i haven't gotten it to work. Trim galore is a wrapper around cutadapt and fastqc to consistently apply adapter and quality trimming to fastq files, with extra functionality for rrbs data. These results are _trimmed.fq files.
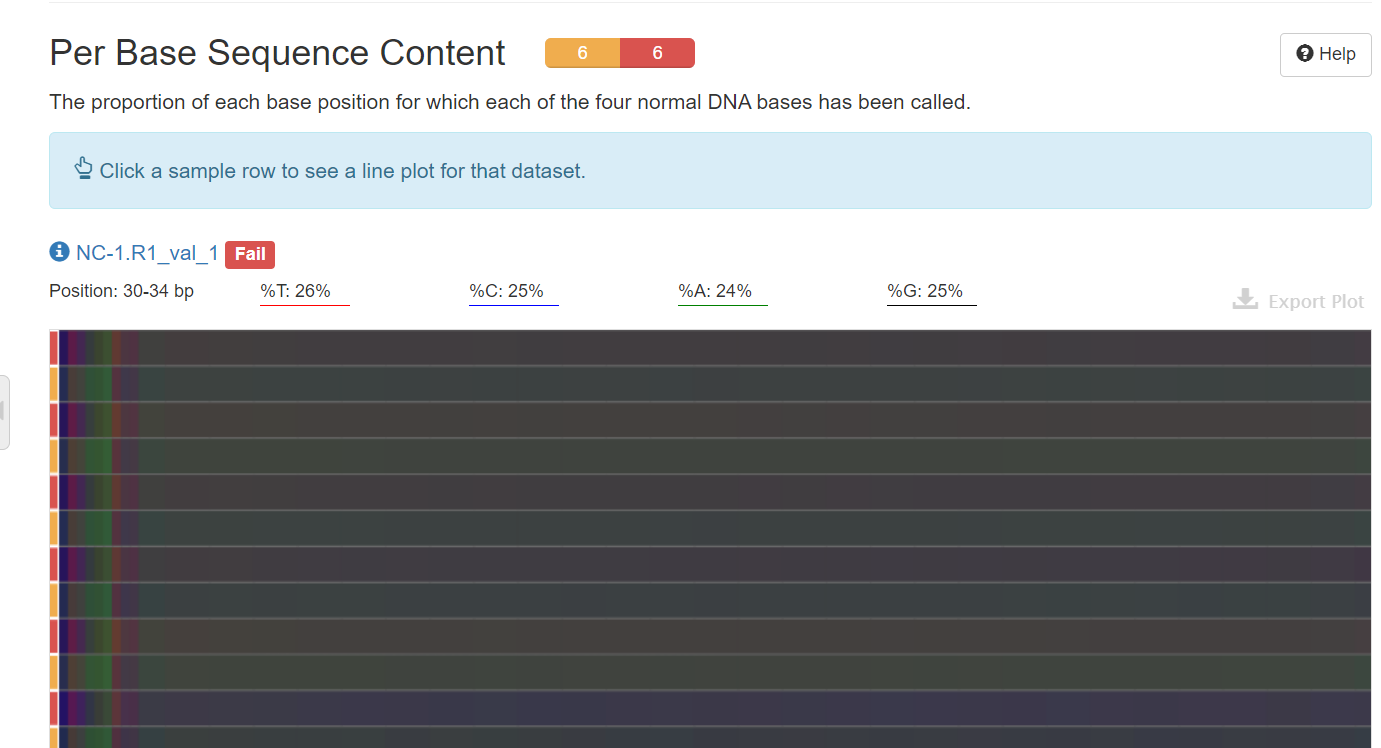
【trim_galore之后的multiqc报告中部分结果还是不理想】_per base sequence content不合格CSDN博客
Trim Galore Biostars The code i'm using is: Is a wrapper script to automate quality and adapter trimming as well as quality control, with some added functionality to. Which tool is the best to trim the raw data? Cutadapt has the parameter setting that asking you to. I'm trying to run trim galore on multiple files (24), but i haven't gotten it to work. As i understand, trim galore! The code i'm using is: Trim galore is a wrapper around cutadapt and fastqc to consistently apply adapter and quality trimming to fastq files, with extra functionality for rrbs data. So some spots may have only one read after. By default, trim galore removes even a single bp match to adapter seq. These results are _trimmed.fq files.
From blog.csdn.net
Trimgalore/fastp对数据的过滤_trimgalore valCSDN博客 Trim Galore Biostars Is a wrapper script to automate quality and adapter trimming as well as quality control, with some added functionality to. Which tool is the best to trim the raw data? By default, trim galore removes even a single bp match to adapter seq. As i understand, trim galore! Trim galore is a wrapper around cutadapt and fastqc to consistently apply. Trim Galore Biostars.
From usermanual.wiki
Trim Galore User Guide V0.4.0 Trim Galore Biostars As i understand, trim galore! Cutadapt has the parameter setting that asking you to. I'm trying to run trim galore on multiple files (24), but i haven't gotten it to work. By default, trim galore removes even a single bp match to adapter seq. The code i'm using is: Is a wrapper script to automate quality and adapter trimming as. Trim Galore Biostars.
From blog.csdn.net
RNAseq——上游分析练习2(数据下载+trimgalore+hisat2+samtools+featureCounts)_上游分析实战 Trim Galore Biostars By default, trim galore removes even a single bp match to adapter seq. So some spots may have only one read after. Trim galore is a wrapper around cutadapt and fastqc to consistently apply adapter and quality trimming to fastq files, with extra functionality for rrbs data. I'm trying to run trim galore on multiple files (24), but i haven't. Trim Galore Biostars.
From blog.csdn.net
RNAseq——上游分析练习2(数据下载+trimgalore+hisat2+samtools+featureCounts)_上游分析实战 Trim Galore Biostars These results are _trimmed.fq files. Is a wrapper script to automate quality and adapter trimming as well as quality control, with some added functionality to. The code i'm using is: Cutadapt has the parameter setting that asking you to. So some spots may have only one read after. By default, trim galore removes even a single bp match to adapter. Trim Galore Biostars.
From 2017-spring-bioinfo.readthedocs.io
Adapter and quality trimming using trimgalore — Inclass Activities Trim Galore Biostars I'm trying to run trim galore on multiple files (24), but i haven't gotten it to work. Which tool is the best to trim the raw data? By default, trim galore removes even a single bp match to adapter seq. These results are _trimmed.fq files. The code i'm using is: Is a wrapper script to automate quality and adapter trimming. Trim Galore Biostars.
From zhuanlan.zhihu.com
详解trim_galore 知乎 Trim Galore Biostars The code i'm using is: These results are _trimmed.fq files. So some spots may have only one read after. Trim galore is a wrapper around cutadapt and fastqc to consistently apply adapter and quality trimming to fastq files, with extra functionality for rrbs data. By default, trim galore removes even a single bp match to adapter seq. Is a wrapper. Trim Galore Biostars.
From phantom-aria.github.io
转录组数据分析笔记(1)——如何用fastqc和trimgalore做测序数据质控 我的小破站 Trim Galore Biostars As i understand, trim galore! Trim galore is a wrapper around cutadapt and fastqc to consistently apply adapter and quality trimming to fastq files, with extra functionality for rrbs data. I'm trying to run trim galore on multiple files (24), but i haven't gotten it to work. The code i'm using is: Which tool is the best to trim the. Trim Galore Biostars.
From phantom-aria.github.io
转录组数据分析笔记(1)——如何用fastqc和trimgalore做测序数据质控 我的小破站 Trim Galore Biostars So some spots may have only one read after. Is a wrapper script to automate quality and adapter trimming as well as quality control, with some added functionality to. These results are _trimmed.fq files. Which tool is the best to trim the raw data? The code i'm using is: Cutadapt has the parameter setting that asking you to. As i. Trim Galore Biostars.
From usermanual.wiki
Trim Galore User Guide V0.4.0 Trim Galore Biostars By default, trim galore removes even a single bp match to adapter seq. The code i'm using is: As i understand, trim galore! So some spots may have only one read after. I'm trying to run trim galore on multiple files (24), but i haven't gotten it to work. Is a wrapper script to automate quality and adapter trimming as. Trim Galore Biostars.
From www.shelven.com
转录组数据分析笔记(1)——如何用fastqc和trimgalore做测序数据质控 我的小破站 Trim Galore Biostars These results are _trimmed.fq files. Trim galore is a wrapper around cutadapt and fastqc to consistently apply adapter and quality trimming to fastq files, with extra functionality for rrbs data. As i understand, trim galore! I'm trying to run trim galore on multiple files (24), but i haven't gotten it to work. Which tool is the best to trim the. Trim Galore Biostars.
From 2017-spring-bioinfo.readthedocs.io
Adapter and quality trimming using trimgalore — Inclass Activities Trim Galore Biostars As i understand, trim galore! These results are _trimmed.fq files. Is a wrapper script to automate quality and adapter trimming as well as quality control, with some added functionality to. I'm trying to run trim galore on multiple files (24), but i haven't gotten it to work. Which tool is the best to trim the raw data? Trim galore is. Trim Galore Biostars.
From blog.csdn.net
RNAseq——上游分析练习2(数据下载+trimgalore+hisat2+samtools+featureCounts)_上游分析实战 Trim Galore Biostars The code i'm using is: Which tool is the best to trim the raw data? Is a wrapper script to automate quality and adapter trimming as well as quality control, with some added functionality to. So some spots may have only one read after. Cutadapt has the parameter setting that asking you to. Trim galore is a wrapper around cutadapt. Trim Galore Biostars.
From www.shelven.com
转录组数据分析笔记(1)——如何用fastqc和trimgalore做测序数据质控 我的小破站 Trim Galore Biostars The code i'm using is: I'm trying to run trim galore on multiple files (24), but i haven't gotten it to work. Which tool is the best to trim the raw data? By default, trim galore removes even a single bp match to adapter seq. Cutadapt has the parameter setting that asking you to. These results are _trimmed.fq files. Trim. Trim Galore Biostars.
From github.com
TrimGalore/Trim_Galore_User_Guide.md at master · FelixKrueger Trim Galore Biostars The code i'm using is: Cutadapt has the parameter setting that asking you to. These results are _trimmed.fq files. I'm trying to run trim galore on multiple files (24), but i haven't gotten it to work. Which tool is the best to trim the raw data? So some spots may have only one read after. Trim galore is a wrapper. Trim Galore Biostars.
From 2017-spring-bioinfo.readthedocs.io
Adapter and quality trimming using trimgalore — Inclass Activities Trim Galore Biostars Trim galore is a wrapper around cutadapt and fastqc to consistently apply adapter and quality trimming to fastq files, with extra functionality for rrbs data. Is a wrapper script to automate quality and adapter trimming as well as quality control, with some added functionality to. So some spots may have only one read after. Cutadapt has the parameter setting that. Trim Galore Biostars.
From blog.csdn.net
Trimgalore/fastp对数据的过滤_trimgalore valCSDN博客 Trim Galore Biostars I'm trying to run trim galore on multiple files (24), but i haven't gotten it to work. Cutadapt has the parameter setting that asking you to. As i understand, trim galore! Trim galore is a wrapper around cutadapt and fastqc to consistently apply adapter and quality trimming to fastq files, with extra functionality for rrbs data. Is a wrapper script. Trim Galore Biostars.
From blog.csdn.net
trim_galore 时,如果没有报错,但结果只有几k 的解决方法Number of sequence pairs removed Trim Galore Biostars Which tool is the best to trim the raw data? The code i'm using is: I'm trying to run trim galore on multiple files (24), but i haven't gotten it to work. Cutadapt has the parameter setting that asking you to. Is a wrapper script to automate quality and adapter trimming as well as quality control, with some added functionality. Trim Galore Biostars.
From gensoft.pasteur.fr
image Trim Galore Biostars So some spots may have only one read after. These results are _trimmed.fq files. Is a wrapper script to automate quality and adapter trimming as well as quality control, with some added functionality to. The code i'm using is: As i understand, trim galore! Which tool is the best to trim the raw data? By default, trim galore removes even. Trim Galore Biostars.
From zhuanlan.zhihu.com
详解trim_galore 知乎 Trim Galore Biostars Which tool is the best to trim the raw data? The code i'm using is: I'm trying to run trim galore on multiple files (24), but i haven't gotten it to work. Is a wrapper script to automate quality and adapter trimming as well as quality control, with some added functionality to. Trim galore is a wrapper around cutadapt and. Trim Galore Biostars.
From blog.csdn.net
Trimgalore/fastp对数据的过滤_trimgalore valCSDN博客 Trim Galore Biostars These results are _trimmed.fq files. So some spots may have only one read after. Is a wrapper script to automate quality and adapter trimming as well as quality control, with some added functionality to. Which tool is the best to trim the raw data? As i understand, trim galore! By default, trim galore removes even a single bp match to. Trim Galore Biostars.
From zhuanlan.zhihu.com
详解trim_galore 知乎 Trim Galore Biostars Cutadapt has the parameter setting that asking you to. Trim galore is a wrapper around cutadapt and fastqc to consistently apply adapter and quality trimming to fastq files, with extra functionality for rrbs data. These results are _trimmed.fq files. The code i'm using is: By default, trim galore removes even a single bp match to adapter seq. So some spots. Trim Galore Biostars.
From blog.csdn.net
RNAseq——上游分析练习2(数据下载+trimgalore+hisat2+samtools+featureCounts)_上游分析实战 Trim Galore Biostars I'm trying to run trim galore on multiple files (24), but i haven't gotten it to work. By default, trim galore removes even a single bp match to adapter seq. Trim galore is a wrapper around cutadapt and fastqc to consistently apply adapter and quality trimming to fastq files, with extra functionality for rrbs data. So some spots may have. Trim Galore Biostars.
From 2017-spring-bioinfo.readthedocs.io
Adapter and quality trimming using trimgalore — Inclass Activities Trim Galore Biostars Trim galore is a wrapper around cutadapt and fastqc to consistently apply adapter and quality trimming to fastq files, with extra functionality for rrbs data. Which tool is the best to trim the raw data? The code i'm using is: As i understand, trim galore! These results are _trimmed.fq files. Cutadapt has the parameter setting that asking you to. By. Trim Galore Biostars.
From gensoft.pasteur.fr
image Trim Galore Biostars These results are _trimmed.fq files. Is a wrapper script to automate quality and adapter trimming as well as quality control, with some added functionality to. The code i'm using is: So some spots may have only one read after. By default, trim galore removes even a single bp match to adapter seq. I'm trying to run trim galore on multiple. Trim Galore Biostars.
From phantom-aria.github.io
转录组数据分析笔记(1)——如何用fastqc和trimgalore做测序数据质控 我的小破站 Trim Galore Biostars Is a wrapper script to automate quality and adapter trimming as well as quality control, with some added functionality to. Which tool is the best to trim the raw data? These results are _trimmed.fq files. Trim galore is a wrapper around cutadapt and fastqc to consistently apply adapter and quality trimming to fastq files, with extra functionality for rrbs data.. Trim Galore Biostars.
From usermanual.wiki
Trim Galore User Guide V0.4.0 Trim Galore Biostars So some spots may have only one read after. I'm trying to run trim galore on multiple files (24), but i haven't gotten it to work. As i understand, trim galore! The code i'm using is: By default, trim galore removes even a single bp match to adapter seq. Is a wrapper script to automate quality and adapter trimming as. Trim Galore Biostars.
From usermanual.wiki
Trim Galore User Guide V0.4.0 Trim Galore Biostars These results are _trimmed.fq files. Is a wrapper script to automate quality and adapter trimming as well as quality control, with some added functionality to. By default, trim galore removes even a single bp match to adapter seq. Which tool is the best to trim the raw data? The code i'm using is: So some spots may have only one. Trim Galore Biostars.
From phantom-aria.github.io
转录组数据分析笔记(1)——如何用fastqc和trimgalore做测序数据质控 我的小破站 Trim Galore Biostars These results are _trimmed.fq files. So some spots may have only one read after. Cutadapt has the parameter setting that asking you to. Trim galore is a wrapper around cutadapt and fastqc to consistently apply adapter and quality trimming to fastq files, with extra functionality for rrbs data. I'm trying to run trim galore on multiple files (24), but i. Trim Galore Biostars.
From 2017-spring-bioinfo.readthedocs.io
Adapter and quality trimming using trimgalore — Inclass Activities Trim Galore Biostars By default, trim galore removes even a single bp match to adapter seq. Is a wrapper script to automate quality and adapter trimming as well as quality control, with some added functionality to. Trim galore is a wrapper around cutadapt and fastqc to consistently apply adapter and quality trimming to fastq files, with extra functionality for rrbs data. The code. Trim Galore Biostars.
From blog.csdn.net
【trim_galore之后的multiqc报告中部分结果还是不理想】_per base sequence content不合格CSDN博客 Trim Galore Biostars I'm trying to run trim galore on multiple files (24), but i haven't gotten it to work. Trim galore is a wrapper around cutadapt and fastqc to consistently apply adapter and quality trimming to fastq files, with extra functionality for rrbs data. Is a wrapper script to automate quality and adapter trimming as well as quality control, with some added. Trim Galore Biostars.
From www.researchgate.net
Workflow of DocMethylPE for WGBS pairedend data analysis, including Trim Galore Biostars Trim galore is a wrapper around cutadapt and fastqc to consistently apply adapter and quality trimming to fastq files, with extra functionality for rrbs data. Is a wrapper script to automate quality and adapter trimming as well as quality control, with some added functionality to. These results are _trimmed.fq files. Cutadapt has the parameter setting that asking you to. The. Trim Galore Biostars.
From blog.csdn.net
RNAseq——上游分析练习2(数据下载+trimgalore+hisat2+samtools+featureCounts)_上游分析实战 Trim Galore Biostars Trim galore is a wrapper around cutadapt and fastqc to consistently apply adapter and quality trimming to fastq files, with extra functionality for rrbs data. These results are _trimmed.fq files. Cutadapt has the parameter setting that asking you to. The code i'm using is: Is a wrapper script to automate quality and adapter trimming as well as quality control, with. Trim Galore Biostars.
From usermanual.wiki
Trim Galore User Guide V0.4.0 Trim Galore Biostars These results are _trimmed.fq files. The code i'm using is: Trim galore is a wrapper around cutadapt and fastqc to consistently apply adapter and quality trimming to fastq files, with extra functionality for rrbs data. By default, trim galore removes even a single bp match to adapter seq. I'm trying to run trim galore on multiple files (24), but i. Trim Galore Biostars.
From blog.csdn.net
【一次性看懂 trim_galore软件的运行代码】_trim galoreCSDN博客 Trim Galore Biostars Is a wrapper script to automate quality and adapter trimming as well as quality control, with some added functionality to. Trim galore is a wrapper around cutadapt and fastqc to consistently apply adapter and quality trimming to fastq files, with extra functionality for rrbs data. So some spots may have only one read after. I'm trying to run trim galore. Trim Galore Biostars.
From usermanual.wiki
Trim Galore User Guide V0.4.0 Trim Galore Biostars So some spots may have only one read after. These results are _trimmed.fq files. By default, trim galore removes even a single bp match to adapter seq. I'm trying to run trim galore on multiple files (24), but i haven't gotten it to work. Trim galore is a wrapper around cutadapt and fastqc to consistently apply adapter and quality trimming. Trim Galore Biostars.