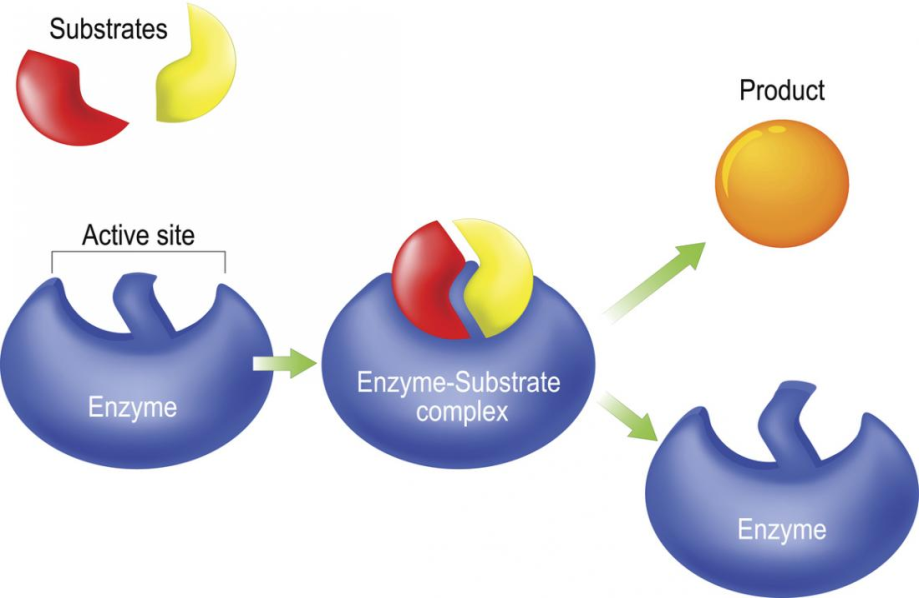
Enzyme Substrate Interaction Models . Enzyme active site and substrate specificity. Here, we introduce viper (virtual interaction predictor for enzyme reactivity), a model that achieves an average 34%. In this study, we combined substrate tolerance data with structural predictions, bioinformatic analysis, molecular. Describe models of substrate binding to an enzyme’s active site. The use of mechanistic modeling of enzyme structure, dynamics, and substrate interactions enables deeper insights into the. The model also assumes that the enzyme is present in a singular, active form, without allosteric modulation or covalent. As the enzyme and substrate come together, their interaction causes a mild shift in the enzyme’s structure that confirms an ideal binding arrangement between the.
from www.w3schools.blog
In this study, we combined substrate tolerance data with structural predictions, bioinformatic analysis, molecular. The use of mechanistic modeling of enzyme structure, dynamics, and substrate interactions enables deeper insights into the. As the enzyme and substrate come together, their interaction causes a mild shift in the enzyme’s structure that confirms an ideal binding arrangement between the. Enzyme active site and substrate specificity. The model also assumes that the enzyme is present in a singular, active form, without allosteric modulation or covalent. Here, we introduce viper (virtual interaction predictor for enzyme reactivity), a model that achieves an average 34%. Describe models of substrate binding to an enzyme’s active site.
Enzymes W3schools
Enzyme Substrate Interaction Models The model also assumes that the enzyme is present in a singular, active form, without allosteric modulation or covalent. The model also assumes that the enzyme is present in a singular, active form, without allosteric modulation or covalent. The use of mechanistic modeling of enzyme structure, dynamics, and substrate interactions enables deeper insights into the. Enzyme active site and substrate specificity. Describe models of substrate binding to an enzyme’s active site. In this study, we combined substrate tolerance data with structural predictions, bioinformatic analysis, molecular. Here, we introduce viper (virtual interaction predictor for enzyme reactivity), a model that achieves an average 34%. As the enzyme and substrate come together, their interaction causes a mild shift in the enzyme’s structure that confirms an ideal binding arrangement between the.
From www.sliderbase.com
Enzymes. A Cell's Catalysts Presentation Biology Enzyme Substrate Interaction Models Here, we introduce viper (virtual interaction predictor for enzyme reactivity), a model that achieves an average 34%. The model also assumes that the enzyme is present in a singular, active form, without allosteric modulation or covalent. As the enzyme and substrate come together, their interaction causes a mild shift in the enzyme’s structure that confirms an ideal binding arrangement between. Enzyme Substrate Interaction Models.
From studymind.co.uk
Enzymes Mechanism of Action (Alevel Biology) Study Mind Enzyme Substrate Interaction Models As the enzyme and substrate come together, their interaction causes a mild shift in the enzyme’s structure that confirms an ideal binding arrangement between the. In this study, we combined substrate tolerance data with structural predictions, bioinformatic analysis, molecular. The use of mechanistic modeling of enzyme structure, dynamics, and substrate interactions enables deeper insights into the. Describe models of substrate. Enzyme Substrate Interaction Models.
From www3.nd.edu
Two Models for EnzymeSubstrate Interactions Enzyme Substrate Interaction Models The model also assumes that the enzyme is present in a singular, active form, without allosteric modulation or covalent. The use of mechanistic modeling of enzyme structure, dynamics, and substrate interactions enables deeper insights into the. Enzyme active site and substrate specificity. Describe models of substrate binding to an enzyme’s active site. In this study, we combined substrate tolerance data. Enzyme Substrate Interaction Models.
From www.slideserve.com
PPT METABOLISME SEL PowerPoint Presentation, free download ID2375676 Enzyme Substrate Interaction Models In this study, we combined substrate tolerance data with structural predictions, bioinformatic analysis, molecular. Here, we introduce viper (virtual interaction predictor for enzyme reactivity), a model that achieves an average 34%. The model also assumes that the enzyme is present in a singular, active form, without allosteric modulation or covalent. Describe models of substrate binding to an enzyme’s active site.. Enzyme Substrate Interaction Models.
From www.slideserve.com
PPT Energy and the Underlying Organization of Life PowerPoint Enzyme Substrate Interaction Models As the enzyme and substrate come together, their interaction causes a mild shift in the enzyme’s structure that confirms an ideal binding arrangement between the. Enzyme active site and substrate specificity. Describe models of substrate binding to an enzyme’s active site. The use of mechanistic modeling of enzyme structure, dynamics, and substrate interactions enables deeper insights into the. The model. Enzyme Substrate Interaction Models.
From mavink.com
What Is An Enzyme Substrate Complex Enzyme Substrate Interaction Models As the enzyme and substrate come together, their interaction causes a mild shift in the enzyme’s structure that confirms an ideal binding arrangement between the. In this study, we combined substrate tolerance data with structural predictions, bioinformatic analysis, molecular. The model also assumes that the enzyme is present in a singular, active form, without allosteric modulation or covalent. Here, we. Enzyme Substrate Interaction Models.
From www.slideserve.com
PPT Chapter 3 Bioenergetics PowerPoint Presentation ID881479 Enzyme Substrate Interaction Models As the enzyme and substrate come together, their interaction causes a mild shift in the enzyme’s structure that confirms an ideal binding arrangement between the. Here, we introduce viper (virtual interaction predictor for enzyme reactivity), a model that achieves an average 34%. In this study, we combined substrate tolerance data with structural predictions, bioinformatic analysis, molecular. The use of mechanistic. Enzyme Substrate Interaction Models.
From animalia-life.club
Enzyme Substrate Enzyme Substrate Interaction Models As the enzyme and substrate come together, their interaction causes a mild shift in the enzyme’s structure that confirms an ideal binding arrangement between the. The model also assumes that the enzyme is present in a singular, active form, without allosteric modulation or covalent. In this study, we combined substrate tolerance data with structural predictions, bioinformatic analysis, molecular. Here, we. Enzyme Substrate Interaction Models.
From www.researchgate.net
Enzymesubstrate interaction modeling strategies. (A) Current machine Enzyme Substrate Interaction Models Here, we introduce viper (virtual interaction predictor for enzyme reactivity), a model that achieves an average 34%. The model also assumes that the enzyme is present in a singular, active form, without allosteric modulation or covalent. Enzyme active site and substrate specificity. In this study, we combined substrate tolerance data with structural predictions, bioinformatic analysis, molecular. The use of mechanistic. Enzyme Substrate Interaction Models.
From www.slideserve.com
PPT Chapter 6 Enzyme PowerPoint Presentation, free download ID5692414 Enzyme Substrate Interaction Models The use of mechanistic modeling of enzyme structure, dynamics, and substrate interactions enables deeper insights into the. As the enzyme and substrate come together, their interaction causes a mild shift in the enzyme’s structure that confirms an ideal binding arrangement between the. Enzyme active site and substrate specificity. Here, we introduce viper (virtual interaction predictor for enzyme reactivity), a model. Enzyme Substrate Interaction Models.
From animalia-life.club
Enzymes Lock And Key Animation Enzyme Substrate Interaction Models The use of mechanistic modeling of enzyme structure, dynamics, and substrate interactions enables deeper insights into the. The model also assumes that the enzyme is present in a singular, active form, without allosteric modulation or covalent. Enzyme active site and substrate specificity. In this study, we combined substrate tolerance data with structural predictions, bioinformatic analysis, molecular. Here, we introduce viper. Enzyme Substrate Interaction Models.
From www.expii.com
Rate of Reaction (Enzymes) — Role & Importance Expii Enzyme Substrate Interaction Models In this study, we combined substrate tolerance data with structural predictions, bioinformatic analysis, molecular. Here, we introduce viper (virtual interaction predictor for enzyme reactivity), a model that achieves an average 34%. The model also assumes that the enzyme is present in a singular, active form, without allosteric modulation or covalent. The use of mechanistic modeling of enzyme structure, dynamics, and. Enzyme Substrate Interaction Models.
From stock.adobe.com
Vecteur Stock Catalysts and enzymes induced fit model. Substrate Enzyme Substrate Interaction Models The use of mechanistic modeling of enzyme structure, dynamics, and substrate interactions enables deeper insights into the. Here, we introduce viper (virtual interaction predictor for enzyme reactivity), a model that achieves an average 34%. In this study, we combined substrate tolerance data with structural predictions, bioinformatic analysis, molecular. Describe models of substrate binding to an enzyme’s active site. Enzyme active. Enzyme Substrate Interaction Models.
From rethinkbiologynotes.com
Enzymesubstrate interaction Rethink Biology Notes Enzyme Substrate Interaction Models The model also assumes that the enzyme is present in a singular, active form, without allosteric modulation or covalent. Here, we introduce viper (virtual interaction predictor for enzyme reactivity), a model that achieves an average 34%. Describe models of substrate binding to an enzyme’s active site. In this study, we combined substrate tolerance data with structural predictions, bioinformatic analysis, molecular.. Enzyme Substrate Interaction Models.
From www.slideserve.com
PPT Ch 4 Energy and Cellular Metabolism PowerPoint Presentation Enzyme Substrate Interaction Models Describe models of substrate binding to an enzyme’s active site. Enzyme active site and substrate specificity. As the enzyme and substrate come together, their interaction causes a mild shift in the enzyme’s structure that confirms an ideal binding arrangement between the. Here, we introduce viper (virtual interaction predictor for enzyme reactivity), a model that achieves an average 34%. The use. Enzyme Substrate Interaction Models.
From www.slideserve.com
PPT General Features of Enzymes PowerPoint Presentation, free Enzyme Substrate Interaction Models The model also assumes that the enzyme is present in a singular, active form, without allosteric modulation or covalent. As the enzyme and substrate come together, their interaction causes a mild shift in the enzyme’s structure that confirms an ideal binding arrangement between the. Here, we introduce viper (virtual interaction predictor for enzyme reactivity), a model that achieves an average. Enzyme Substrate Interaction Models.
From www.youtube.com
Enzyme Substrate Interaction = Lock and Key Concept of Enzyme Lock Enzyme Substrate Interaction Models Enzyme active site and substrate specificity. As the enzyme and substrate come together, their interaction causes a mild shift in the enzyme’s structure that confirms an ideal binding arrangement between the. In this study, we combined substrate tolerance data with structural predictions, bioinformatic analysis, molecular. The model also assumes that the enzyme is present in a singular, active form, without. Enzyme Substrate Interaction Models.
From www.slideserve.com
PPT Chapter 13 Introduction to Enzymes PowerPoint Presentation, free Enzyme Substrate Interaction Models The model also assumes that the enzyme is present in a singular, active form, without allosteric modulation or covalent. Enzyme active site and substrate specificity. In this study, we combined substrate tolerance data with structural predictions, bioinformatic analysis, molecular. The use of mechanistic modeling of enzyme structure, dynamics, and substrate interactions enables deeper insights into the. As the enzyme and. Enzyme Substrate Interaction Models.
From www.numerade.com
SOLVED The induced fit model of enzymesubstrate interaction proposes Enzyme Substrate Interaction Models Describe models of substrate binding to an enzyme’s active site. As the enzyme and substrate come together, their interaction causes a mild shift in the enzyme’s structure that confirms an ideal binding arrangement between the. The model also assumes that the enzyme is present in a singular, active form, without allosteric modulation or covalent. Enzyme active site and substrate specificity.. Enzyme Substrate Interaction Models.
From www.dreamstime.com
Model of Enzyme Activity Induced Fir Model of Catalytic Action with Enzyme Substrate Interaction Models In this study, we combined substrate tolerance data with structural predictions, bioinformatic analysis, molecular. Here, we introduce viper (virtual interaction predictor for enzyme reactivity), a model that achieves an average 34%. Describe models of substrate binding to an enzyme’s active site. Enzyme active site and substrate specificity. As the enzyme and substrate come together, their interaction causes a mild shift. Enzyme Substrate Interaction Models.
From www.biologyonline.com
Substrate Definition and Examples Biology Online Dictionary Enzyme Substrate Interaction Models Here, we introduce viper (virtual interaction predictor for enzyme reactivity), a model that achieves an average 34%. As the enzyme and substrate come together, their interaction causes a mild shift in the enzyme’s structure that confirms an ideal binding arrangement between the. In this study, we combined substrate tolerance data with structural predictions, bioinformatic analysis, molecular. The use of mechanistic. Enzyme Substrate Interaction Models.
From www.slideserve.com
PPT Enzyme Mechanisms PowerPoint Presentation, free download ID3783127 Enzyme Substrate Interaction Models In this study, we combined substrate tolerance data with structural predictions, bioinformatic analysis, molecular. As the enzyme and substrate come together, their interaction causes a mild shift in the enzyme’s structure that confirms an ideal binding arrangement between the. Enzyme active site and substrate specificity. The model also assumes that the enzyme is present in a singular, active form, without. Enzyme Substrate Interaction Models.
From www.alamy.com
lock and key model of enzyme, Biological model of lock and key Enzyme Substrate Interaction Models Describe models of substrate binding to an enzyme’s active site. In this study, we combined substrate tolerance data with structural predictions, bioinformatic analysis, molecular. Enzyme active site and substrate specificity. The use of mechanistic modeling of enzyme structure, dynamics, and substrate interactions enables deeper insights into the. The model also assumes that the enzyme is present in a singular, active. Enzyme Substrate Interaction Models.
From science.halleyhosting.com
Chapter 8 Enzymes Enzyme Substrate Interaction Models Here, we introduce viper (virtual interaction predictor for enzyme reactivity), a model that achieves an average 34%. The use of mechanistic modeling of enzyme structure, dynamics, and substrate interactions enables deeper insights into the. The model also assumes that the enzyme is present in a singular, active form, without allosteric modulation or covalent. As the enzyme and substrate come together,. Enzyme Substrate Interaction Models.
From ibiologia.com
Substrate Definition , Biochemsitry & Examples Enzyme Substrate Interaction Models Enzyme active site and substrate specificity. As the enzyme and substrate come together, their interaction causes a mild shift in the enzyme’s structure that confirms an ideal binding arrangement between the. The model also assumes that the enzyme is present in a singular, active form, without allosteric modulation or covalent. Describe models of substrate binding to an enzyme’s active site.. Enzyme Substrate Interaction Models.
From biology4ibdp.weebly.com
2.5 Enzymes BIOLOGY4IBDP Enzyme Substrate Interaction Models Describe models of substrate binding to an enzyme’s active site. Here, we introduce viper (virtual interaction predictor for enzyme reactivity), a model that achieves an average 34%. The use of mechanistic modeling of enzyme structure, dynamics, and substrate interactions enables deeper insights into the. As the enzyme and substrate come together, their interaction causes a mild shift in the enzyme’s. Enzyme Substrate Interaction Models.
From www.researchgate.net
Effects of enzymesubstrate interactions on the free energy difference Enzyme Substrate Interaction Models The use of mechanistic modeling of enzyme structure, dynamics, and substrate interactions enables deeper insights into the. Describe models of substrate binding to an enzyme’s active site. Enzyme active site and substrate specificity. As the enzyme and substrate come together, their interaction causes a mild shift in the enzyme’s structure that confirms an ideal binding arrangement between the. In this. Enzyme Substrate Interaction Models.
From stock.adobe.com
Biological diagram show mechanism of enzyme substrate interaction by Enzyme Substrate Interaction Models Describe models of substrate binding to an enzyme’s active site. The use of mechanistic modeling of enzyme structure, dynamics, and substrate interactions enables deeper insights into the. As the enzyme and substrate come together, their interaction causes a mild shift in the enzyme’s structure that confirms an ideal binding arrangement between the. Here, we introduce viper (virtual interaction predictor for. Enzyme Substrate Interaction Models.
From www.w3schools.blog
Enzymes W3schools Enzyme Substrate Interaction Models As the enzyme and substrate come together, their interaction causes a mild shift in the enzyme’s structure that confirms an ideal binding arrangement between the. In this study, we combined substrate tolerance data with structural predictions, bioinformatic analysis, molecular. The model also assumes that the enzyme is present in a singular, active form, without allosteric modulation or covalent. Here, we. Enzyme Substrate Interaction Models.
From kabardesa.my.id
Biological diagram show mechanism of enzyme substrate interaction by Enzyme Substrate Interaction Models In this study, we combined substrate tolerance data with structural predictions, bioinformatic analysis, molecular. Describe models of substrate binding to an enzyme’s active site. Enzyme active site and substrate specificity. The use of mechanistic modeling of enzyme structure, dynamics, and substrate interactions enables deeper insights into the. The model also assumes that the enzyme is present in a singular, active. Enzyme Substrate Interaction Models.
From psiberg.com
What affects enzyme activity? Biochemistry PSIBERG Enzyme Substrate Interaction Models Enzyme active site and substrate specificity. The model also assumes that the enzyme is present in a singular, active form, without allosteric modulation or covalent. In this study, we combined substrate tolerance data with structural predictions, bioinformatic analysis, molecular. Here, we introduce viper (virtual interaction predictor for enzyme reactivity), a model that achieves an average 34%. As the enzyme and. Enzyme Substrate Interaction Models.
From www.sliderbase.com
Enzymes. A Cell's Catalysts Presentation Biology Enzyme Substrate Interaction Models Describe models of substrate binding to an enzyme’s active site. Here, we introduce viper (virtual interaction predictor for enzyme reactivity), a model that achieves an average 34%. As the enzyme and substrate come together, their interaction causes a mild shift in the enzyme’s structure that confirms an ideal binding arrangement between the. Enzyme active site and substrate specificity. In this. Enzyme Substrate Interaction Models.
From slideserve.com
PPT Protein Structure and Enzyme Function PowerPoint Presentation Enzyme Substrate Interaction Models The model also assumes that the enzyme is present in a singular, active form, without allosteric modulation or covalent. The use of mechanistic modeling of enzyme structure, dynamics, and substrate interactions enables deeper insights into the. In this study, we combined substrate tolerance data with structural predictions, bioinformatic analysis, molecular. Enzyme active site and substrate specificity. Here, we introduce viper. Enzyme Substrate Interaction Models.
From www.alamy.com
The combination formed by an enzyme and its substrates is called the Enzyme Substrate Interaction Models Describe models of substrate binding to an enzyme’s active site. Here, we introduce viper (virtual interaction predictor for enzyme reactivity), a model that achieves an average 34%. The model also assumes that the enzyme is present in a singular, active form, without allosteric modulation or covalent. The use of mechanistic modeling of enzyme structure, dynamics, and substrate interactions enables deeper. Enzyme Substrate Interaction Models.
From www.researchgate.net
2D view of enzymesubstrate interactions. The hydrogen bonds were Enzyme Substrate Interaction Models In this study, we combined substrate tolerance data with structural predictions, bioinformatic analysis, molecular. The use of mechanistic modeling of enzyme structure, dynamics, and substrate interactions enables deeper insights into the. Enzyme active site and substrate specificity. As the enzyme and substrate come together, their interaction causes a mild shift in the enzyme’s structure that confirms an ideal binding arrangement. Enzyme Substrate Interaction Models.