Protein-Protein Blast . Blastx compares a nucleotide query sequence to a protein sequence database by. Blastp compares a protein query sequence to a protein sequence database. Align two or more protein sequences with clustal omega to find conserved regions The program compares nucleotide or protein sequences to sequence. Blast 1 is widely used in molecular biology to search for nucleotide and protein sequences. The basic local alignment search tool (blast) finds regions of local similarity between sequences. Select the blast tab of the toolbar to run a sequence similarity search with the blast (basic local alignment search tool) program:
from abacus.bates.edu
The basic local alignment search tool (blast) finds regions of local similarity between sequences. Align two or more protein sequences with clustal omega to find conserved regions Blastp compares a protein query sequence to a protein sequence database. The program compares nucleotide or protein sequences to sequence. Blast 1 is widely used in molecular biology to search for nucleotide and protein sequences. Select the blast tab of the toolbar to run a sequence similarity search with the blast (basic local alignment search tool) program: Blastx compares a nucleotide query sequence to a protein sequence database by.
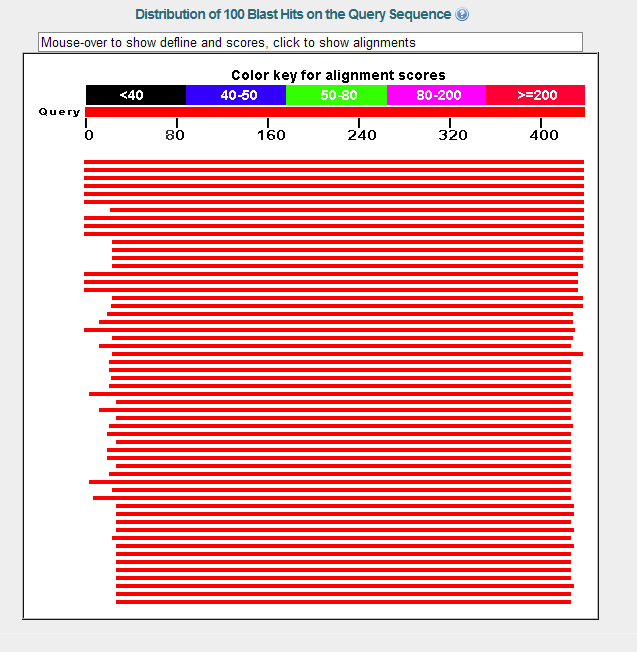
BLASTP Results Part 1
Protein-Protein Blast Blastp compares a protein query sequence to a protein sequence database. Blastp compares a protein query sequence to a protein sequence database. Align two or more protein sequences with clustal omega to find conserved regions Blast 1 is widely used in molecular biology to search for nucleotide and protein sequences. Blastx compares a nucleotide query sequence to a protein sequence database by. Select the blast tab of the toolbar to run a sequence similarity search with the blast (basic local alignment search tool) program: The program compares nucleotide or protein sequences to sequence. The basic local alignment search tool (blast) finds regions of local similarity between sequences.
From www.slideserve.com
PPT BLAST PowerPoint Presentation, free download ID3384708 Protein-Protein Blast Blast 1 is widely used in molecular biology to search for nucleotide and protein sequences. The basic local alignment search tool (blast) finds regions of local similarity between sequences. Select the blast tab of the toolbar to run a sequence similarity search with the blast (basic local alignment search tool) program: Blastx compares a nucleotide query sequence to a protein. Protein-Protein Blast.
From digitalworldbiology.com
Using protein blast and searching for Elvis, part I. Digital World Protein-Protein Blast Blastp compares a protein query sequence to a protein sequence database. Blast 1 is widely used in molecular biology to search for nucleotide and protein sequences. Align two or more protein sequences with clustal omega to find conserved regions The basic local alignment search tool (blast) finds regions of local similarity between sequences. The program compares nucleotide or protein sequences. Protein-Protein Blast.
From www.walmart.ca
BioX All Natural Protein Blast Caramel Fudge Protein bar Walmart Canada Protein-Protein Blast The basic local alignment search tool (blast) finds regions of local similarity between sequences. The program compares nucleotide or protein sequences to sequence. Select the blast tab of the toolbar to run a sequence similarity search with the blast (basic local alignment search tool) program: Blastp compares a protein query sequence to a protein sequence database. Align two or more. Protein-Protein Blast.
From fity.club
Blastn Protein-Protein Blast Blastp compares a protein query sequence to a protein sequence database. Select the blast tab of the toolbar to run a sequence similarity search with the blast (basic local alignment search tool) program: The program compares nucleotide or protein sequences to sequence. The basic local alignment search tool (blast) finds regions of local similarity between sequences. Blast 1 is widely. Protein-Protein Blast.
From www.slideserve.com
PPT Gapped BLAST and PSIBLAST : a new generation of protein database Protein-Protein Blast Blast 1 is widely used in molecular biology to search for nucleotide and protein sequences. Select the blast tab of the toolbar to run a sequence similarity search with the blast (basic local alignment search tool) program: Blastp compares a protein query sequence to a protein sequence database. The program compares nucleotide or protein sequences to sequence. Blastx compares a. Protein-Protein Blast.
From ljseedco.com
HEMPAGIZER PROTEIN BLAST 1lb Lady Jane Gourmet Seed Co Protein-Protein Blast The program compares nucleotide or protein sequences to sequence. Select the blast tab of the toolbar to run a sequence similarity search with the blast (basic local alignment search tool) program: Align two or more protein sequences with clustal omega to find conserved regions Blast 1 is widely used in molecular biology to search for nucleotide and protein sequences. The. Protein-Protein Blast.
From www.londondrugs.com
BIOX PROTEIN BLAST BAR C/FUD 72G Protein-Protein Blast The basic local alignment search tool (blast) finds regions of local similarity between sequences. Blastx compares a nucleotide query sequence to a protein sequence database by. Align two or more protein sequences with clustal omega to find conserved regions The program compares nucleotide or protein sequences to sequence. Blast 1 is widely used in molecular biology to search for nucleotide. Protein-Protein Blast.
From present5.com
Classifying the protein universe Synapse Associated Protein 97 Protein-Protein Blast Blastp compares a protein query sequence to a protein sequence database. Blast 1 is widely used in molecular biology to search for nucleotide and protein sequences. The program compares nucleotide or protein sequences to sequence. Blastx compares a nucleotide query sequence to a protein sequence database by. Align two or more protein sequences with clustal omega to find conserved regions. Protein-Protein Blast.
From www.slideserve.com
PPT NCBI Molecular Biology Resources PowerPoint Presentation, free Protein-Protein Blast The basic local alignment search tool (blast) finds regions of local similarity between sequences. Blastx compares a nucleotide query sequence to a protein sequence database by. The program compares nucleotide or protein sequences to sequence. Select the blast tab of the toolbar to run a sequence similarity search with the blast (basic local alignment search tool) program: Blastp compares a. Protein-Protein Blast.
From www.youtube.com
Which Database to Choose for Protein BLAST run YouTube Protein-Protein Blast Blast 1 is widely used in molecular biology to search for nucleotide and protein sequences. The program compares nucleotide or protein sequences to sequence. The basic local alignment search tool (blast) finds regions of local similarity between sequences. Blastp compares a protein query sequence to a protein sequence database. Select the blast tab of the toolbar to run a sequence. Protein-Protein Blast.
From joibnsqro.blob.core.windows.net
Functional Protein Definition Biology at Leona Jackson blog Protein-Protein Blast Align two or more protein sequences with clustal omega to find conserved regions Blastp compares a protein query sequence to a protein sequence database. The program compares nucleotide or protein sequences to sequence. The basic local alignment search tool (blast) finds regions of local similarity between sequences. Blastx compares a nucleotide query sequence to a protein sequence database by. Select. Protein-Protein Blast.
From ljseedco.com
HEMPAGIZER PROTEIN BLAST 1lb Lady Jane Gourmet Seed Co Protein-Protein Blast The basic local alignment search tool (blast) finds regions of local similarity between sequences. The program compares nucleotide or protein sequences to sequence. Align two or more protein sequences with clustal omega to find conserved regions Select the blast tab of the toolbar to run a sequence similarity search with the blast (basic local alignment search tool) program: Blastp compares. Protein-Protein Blast.
From www.slideserve.com
PPT CED9 PowerPoint Presentation, free download ID3429166 Protein-Protein Blast Blastp compares a protein query sequence to a protein sequence database. Blastx compares a nucleotide query sequence to a protein sequence database by. Blast 1 is widely used in molecular biology to search for nucleotide and protein sequences. Align two or more protein sequences with clustal omega to find conserved regions The program compares nucleotide or protein sequences to sequence.. Protein-Protein Blast.
From www.youtube.com
Which BLAST Algorithm to use for ProteinProtein BLAST YouTube Protein-Protein Blast The basic local alignment search tool (blast) finds regions of local similarity between sequences. Align two or more protein sequences with clustal omega to find conserved regions Blastp compares a protein query sequence to a protein sequence database. Select the blast tab of the toolbar to run a sequence similarity search with the blast (basic local alignment search tool) program:. Protein-Protein Blast.
From fity.club
Nucleotide Blast Protein-Protein Blast Blastp compares a protein query sequence to a protein sequence database. Blastx compares a nucleotide query sequence to a protein sequence database by. Select the blast tab of the toolbar to run a sequence similarity search with the blast (basic local alignment search tool) program: Align two or more protein sequences with clustal omega to find conserved regions Blast 1. Protein-Protein Blast.
From www.walmart.ca
BioX All Natural Protein Blast Creamy Peanut Fudge Protein Bar Protein-Protein Blast Blastp compares a protein query sequence to a protein sequence database. Align two or more protein sequences with clustal omega to find conserved regions Select the blast tab of the toolbar to run a sequence similarity search with the blast (basic local alignment search tool) program: Blast 1 is widely used in molecular biology to search for nucleotide and protein. Protein-Protein Blast.
From www.wholefoodsmarket.com
Recipe Whey Protein Breakfast Blast Whole Foods Market Protein-Protein Blast Blast 1 is widely used in molecular biology to search for nucleotide and protein sequences. The basic local alignment search tool (blast) finds regions of local similarity between sequences. Blastx compares a nucleotide query sequence to a protein sequence database by. The program compares nucleotide or protein sequences to sequence. Blastp compares a protein query sequence to a protein sequence. Protein-Protein Blast.
From www.researchgate.net
Species distribution of the standard proteinprotein BLAST (BLASTp Protein-Protein Blast Blast 1 is widely used in molecular biology to search for nucleotide and protein sequences. Align two or more protein sequences with clustal omega to find conserved regions The basic local alignment search tool (blast) finds regions of local similarity between sequences. Blastp compares a protein query sequence to a protein sequence database. The program compares nucleotide or protein sequences. Protein-Protein Blast.
From www.slideserve.com
PPT Protein Sequence PowerPoint Presentation, free download ID4271810 Protein-Protein Blast Align two or more protein sequences with clustal omega to find conserved regions The program compares nucleotide or protein sequences to sequence. Blast 1 is widely used in molecular biology to search for nucleotide and protein sequences. Blastp compares a protein query sequence to a protein sequence database. Blastx compares a nucleotide query sequence to a protein sequence database by.. Protein-Protein Blast.
From www.mdpi.com
Life Free FullText NonStandard Codes Define New Concepts Protein-Protein Blast Blastx compares a nucleotide query sequence to a protein sequence database by. Align two or more protein sequences with clustal omega to find conserved regions Select the blast tab of the toolbar to run a sequence similarity search with the blast (basic local alignment search tool) program: The program compares nucleotide or protein sequences to sequence. Blast 1 is widely. Protein-Protein Blast.
From chocolatierindore.in
Protein Blast Chocolatier Protein-Protein Blast Blast 1 is widely used in molecular biology to search for nucleotide and protein sequences. The program compares nucleotide or protein sequences to sequence. Blastx compares a nucleotide query sequence to a protein sequence database by. Blastp compares a protein query sequence to a protein sequence database. Select the blast tab of the toolbar to run a sequence similarity search. Protein-Protein Blast.
From www.psb.ugent.be
PTM Viewer Protein-Protein Blast Blast 1 is widely used in molecular biology to search for nucleotide and protein sequences. The program compares nucleotide or protein sequences to sequence. The basic local alignment search tool (blast) finds regions of local similarity between sequences. Blastp compares a protein query sequence to a protein sequence database. Blastx compares a nucleotide query sequence to a protein sequence database. Protein-Protein Blast.
From abacus.bates.edu
BLASTP Results Part 1 Protein-Protein Blast The basic local alignment search tool (blast) finds regions of local similarity between sequences. The program compares nucleotide or protein sequences to sequence. Align two or more protein sequences with clustal omega to find conserved regions Blastx compares a nucleotide query sequence to a protein sequence database by. Select the blast tab of the toolbar to run a sequence similarity. Protein-Protein Blast.
From www.researchgate.net
Protein structure prediction pipeline. The protein structure prediction Protein-Protein Blast Blastx compares a nucleotide query sequence to a protein sequence database by. Blast 1 is widely used in molecular biology to search for nucleotide and protein sequences. The basic local alignment search tool (blast) finds regions of local similarity between sequences. Align two or more protein sequences with clustal omega to find conserved regions The program compares nucleotide or protein. Protein-Protein Blast.
From www.slideserve.com
PPT Using and Analyzing Fluorescent Proteins BLAST KEY PowerPoint Protein-Protein Blast Align two or more protein sequences with clustal omega to find conserved regions Blastx compares a nucleotide query sequence to a protein sequence database by. The basic local alignment search tool (blast) finds regions of local similarity between sequences. The program compares nucleotide or protein sequences to sequence. Blast 1 is widely used in molecular biology to search for nucleotide. Protein-Protein Blast.
From www.youtube.com
bioinformatics step by step guide how to use protein blast in NCBI Protein-Protein Blast The basic local alignment search tool (blast) finds regions of local similarity between sequences. Select the blast tab of the toolbar to run a sequence similarity search with the blast (basic local alignment search tool) program: Blastp compares a protein query sequence to a protein sequence database. Align two or more protein sequences with clustal omega to find conserved regions. Protein-Protein Blast.
From zonenutrition.me
Biox, All Natural Protein Blast Bar, (12/Box) Zone Nutrition Protein-Protein Blast Blastx compares a nucleotide query sequence to a protein sequence database by. Align two or more protein sequences with clustal omega to find conserved regions Blast 1 is widely used in molecular biology to search for nucleotide and protein sequences. Select the blast tab of the toolbar to run a sequence similarity search with the blast (basic local alignment search. Protein-Protein Blast.
From www.researchgate.net
NCBI Protein BLAST Taxonomy OmpC321340 Download Scientific Diagram Protein-Protein Blast Blastp compares a protein query sequence to a protein sequence database. Blast 1 is widely used in molecular biology to search for nucleotide and protein sequences. The basic local alignment search tool (blast) finds regions of local similarity between sequences. Align two or more protein sequences with clustal omega to find conserved regions Blastx compares a nucleotide query sequence to. Protein-Protein Blast.
From www.slideserve.com
PPT BLAST PowerPoint Presentation, free download ID3384708 Protein-Protein Blast Blast 1 is widely used in molecular biology to search for nucleotide and protein sequences. Align two or more protein sequences with clustal omega to find conserved regions Blastx compares a nucleotide query sequence to a protein sequence database by. Blastp compares a protein query sequence to a protein sequence database. The program compares nucleotide or protein sequences to sequence.. Protein-Protein Blast.
From www.slideserve.com
PPT Introduction to BLAST PowerPoint Presentation, free download ID Protein-Protein Blast Blastx compares a nucleotide query sequence to a protein sequence database by. Blastp compares a protein query sequence to a protein sequence database. Align two or more protein sequences with clustal omega to find conserved regions Blast 1 is widely used in molecular biology to search for nucleotide and protein sequences. Select the blast tab of the toolbar to run. Protein-Protein Blast.
From www.slideserve.com
PPT Diversity of Life Introduction to Biological Classification Protein-Protein Blast The basic local alignment search tool (blast) finds regions of local similarity between sequences. Align two or more protein sequences with clustal omega to find conserved regions Select the blast tab of the toolbar to run a sequence similarity search with the blast (basic local alignment search tool) program: The program compares nucleotide or protein sequences to sequence. Blastx compares. Protein-Protein Blast.
From www.londondrugs.com
BIOX PROTEIN BLAST BAR PEANU 72G Protein-Protein Blast Blastx compares a nucleotide query sequence to a protein sequence database by. Align two or more protein sequences with clustal omega to find conserved regions Select the blast tab of the toolbar to run a sequence similarity search with the blast (basic local alignment search tool) program: The program compares nucleotide or protein sequences to sequence. Blastp compares a protein. Protein-Protein Blast.
From widdowquinn.github.io
01blast_at_NCBI_website Protein-Protein Blast Align two or more protein sequences with clustal omega to find conserved regions Blastp compares a protein query sequence to a protein sequence database. The program compares nucleotide or protein sequences to sequence. The basic local alignment search tool (blast) finds regions of local similarity between sequences. Blast 1 is widely used in molecular biology to search for nucleotide and. Protein-Protein Blast.
From www.slideserve.com
PPT CED9 PowerPoint Presentation, free download ID3429166 Protein-Protein Blast Blast 1 is widely used in molecular biology to search for nucleotide and protein sequences. Blastp compares a protein query sequence to a protein sequence database. The basic local alignment search tool (blast) finds regions of local similarity between sequences. Blastx compares a nucleotide query sequence to a protein sequence database by. The program compares nucleotide or protein sequences to. Protein-Protein Blast.
From www.youtube.com
Translated Nucleotide to Protein BLAST BLASTX YouTube Protein-Protein Blast Blast 1 is widely used in molecular biology to search for nucleotide and protein sequences. The program compares nucleotide or protein sequences to sequence. Blastx compares a nucleotide query sequence to a protein sequence database by. Select the blast tab of the toolbar to run a sequence similarity search with the blast (basic local alignment search tool) program: Blastp compares. Protein-Protein Blast.