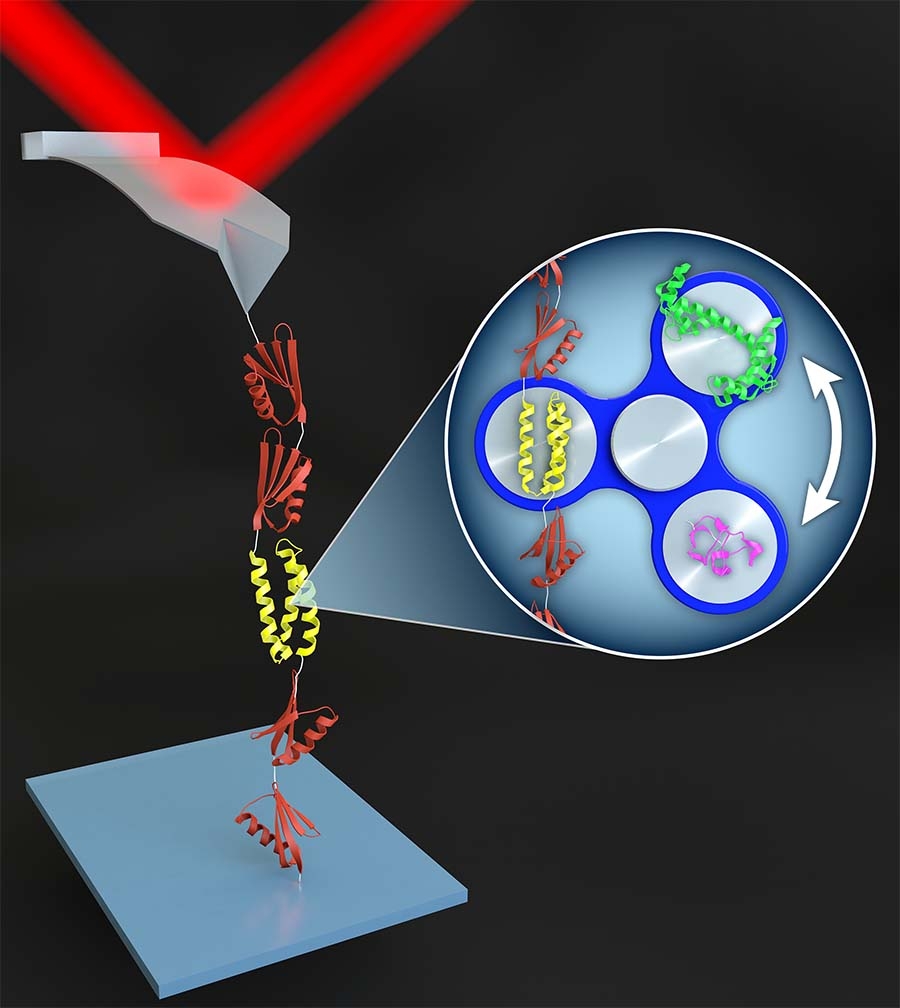
Jacs Protein Folding . Coli cells for two protein gb3 mutants. Decades of research on protein folding have primarily. — in this work, we measured the millisecond residue specific protein folding and unfolding dynamics in e. — proteins g and l fold to similar topologies despite low sequence similarity, but differ in their folding pathways. The principle is based on constructing a.
from jila.colorado.edu
— in this work, we measured the millisecond residue specific protein folding and unfolding dynamics in e. The principle is based on constructing a. Coli cells for two protein gb3 mutants. — proteins g and l fold to similar topologies despite low sequence similarity, but differ in their folding pathways. Decades of research on protein folding have primarily.
Protein folding and mechanobiology JILA Exploring the Frontiers of
Jacs Protein Folding Coli cells for two protein gb3 mutants. The principle is based on constructing a. Decades of research on protein folding have primarily. Coli cells for two protein gb3 mutants. — proteins g and l fold to similar topologies despite low sequence similarity, but differ in their folding pathways. — in this work, we measured the millisecond residue specific protein folding and unfolding dynamics in e.
From achs-prod.acs.org
Mechanism of Coupled FoldinguponBinding of an Intrinsically Jacs Protein Folding Coli cells for two protein gb3 mutants. The principle is based on constructing a. — proteins g and l fold to similar topologies despite low sequence similarity, but differ in their folding pathways. Decades of research on protein folding have primarily. — in this work, we measured the millisecond residue specific protein folding and unfolding dynamics in e. Jacs Protein Folding.
From pubs.acs.org
MechanismBased Strategy for Optimizing HaloTag Protein Labeling JACS Au Jacs Protein Folding — in this work, we measured the millisecond residue specific protein folding and unfolding dynamics in e. Coli cells for two protein gb3 mutants. Decades of research on protein folding have primarily. The principle is based on constructing a. — proteins g and l fold to similar topologies despite low sequence similarity, but differ in their folding pathways. Jacs Protein Folding.
From www.researchgate.net
3 Models for protein folding (reprinted from (Nolting and Andert Jacs Protein Folding The principle is based on constructing a. Decades of research on protein folding have primarily. — in this work, we measured the millisecond residue specific protein folding and unfolding dynamics in e. Coli cells for two protein gb3 mutants. — proteins g and l fold to similar topologies despite low sequence similarity, but differ in their folding pathways. Jacs Protein Folding.
From pubs.acs.org
Secondary Structural Change Can Occur Diffusely and Not Modularly Jacs Protein Folding — in this work, we measured the millisecond residue specific protein folding and unfolding dynamics in e. — proteins g and l fold to similar topologies despite low sequence similarity, but differ in their folding pathways. The principle is based on constructing a. Decades of research on protein folding have primarily. Coli cells for two protein gb3 mutants. Jacs Protein Folding.
From www.biorxiv.org
Tailored Design of Protein Nanoparticle Scaffolds for Multivalent Jacs Protein Folding Decades of research on protein folding have primarily. — in this work, we measured the millisecond residue specific protein folding and unfolding dynamics in e. Coli cells for two protein gb3 mutants. The principle is based on constructing a. — proteins g and l fold to similar topologies despite low sequence similarity, but differ in their folding pathways. Jacs Protein Folding.
From pubs.acs.org
Structural Characterization of the Early Events in the Nucleation Jacs Protein Folding Decades of research on protein folding have primarily. — proteins g and l fold to similar topologies despite low sequence similarity, but differ in their folding pathways. The principle is based on constructing a. — in this work, we measured the millisecond residue specific protein folding and unfolding dynamics in e. Coli cells for two protein gb3 mutants. Jacs Protein Folding.
From pubs.acs.org
Deciphering the Folding Mechanism of Proteins G and L and Their Mutants Jacs Protein Folding Coli cells for two protein gb3 mutants. Decades of research on protein folding have primarily. The principle is based on constructing a. — in this work, we measured the millisecond residue specific protein folding and unfolding dynamics in e. — proteins g and l fold to similar topologies despite low sequence similarity, but differ in their folding pathways. Jacs Protein Folding.
From mavink.com
Protein Folding Pathway Jacs Protein Folding — proteins g and l fold to similar topologies despite low sequence similarity, but differ in their folding pathways. Coli cells for two protein gb3 mutants. — in this work, we measured the millisecond residue specific protein folding and unfolding dynamics in e. Decades of research on protein folding have primarily. The principle is based on constructing a. Jacs Protein Folding.
From dokumen.tips
(PDF) Protein folding SimBacThe folding mechanism A polypeptide Jacs Protein Folding The principle is based on constructing a. Coli cells for two protein gb3 mutants. — in this work, we measured the millisecond residue specific protein folding and unfolding dynamics in e. Decades of research on protein folding have primarily. — proteins g and l fold to similar topologies despite low sequence similarity, but differ in their folding pathways. Jacs Protein Folding.
From achs-prod.acs.org
Interaction Networks in Protein Folding via AtomicResolution Jacs Protein Folding — in this work, we measured the millisecond residue specific protein folding and unfolding dynamics in e. — proteins g and l fold to similar topologies despite low sequence similarity, but differ in their folding pathways. Coli cells for two protein gb3 mutants. Decades of research on protein folding have primarily. The principle is based on constructing a. Jacs Protein Folding.
From testpubschina.acs.org
High Pressure ZZExchange NMR Reveals Key Features of Protein Folding Jacs Protein Folding — in this work, we measured the millisecond residue specific protein folding and unfolding dynamics in e. — proteins g and l fold to similar topologies despite low sequence similarity, but differ in their folding pathways. The principle is based on constructing a. Decades of research on protein folding have primarily. Coli cells for two protein gb3 mutants. Jacs Protein Folding.
From achs-prod.acs.org
Physical Origins of Codon Positions That Strongly Influence Jacs Protein Folding The principle is based on constructing a. — in this work, we measured the millisecond residue specific protein folding and unfolding dynamics in e. Coli cells for two protein gb3 mutants. — proteins g and l fold to similar topologies despite low sequence similarity, but differ in their folding pathways. Decades of research on protein folding have primarily. Jacs Protein Folding.
From pubs.acs.org
Deep Learning Dynamic Allostery of GProteinCoupled Receptors JACS Au Jacs Protein Folding — proteins g and l fold to similar topologies despite low sequence similarity, but differ in their folding pathways. The principle is based on constructing a. Decades of research on protein folding have primarily. — in this work, we measured the millisecond residue specific protein folding and unfolding dynamics in e. Coli cells for two protein gb3 mutants. Jacs Protein Folding.
From pubs.acs.org
FoldingDegradation Relationship of a Membrane Protein Mediated by the Jacs Protein Folding Decades of research on protein folding have primarily. — proteins g and l fold to similar topologies despite low sequence similarity, but differ in their folding pathways. The principle is based on constructing a. Coli cells for two protein gb3 mutants. — in this work, we measured the millisecond residue specific protein folding and unfolding dynamics in e. Jacs Protein Folding.
From dokumen.tips
(PDF) New Protein Folding Discrete Molecular Dynamics Studybrigita Jacs Protein Folding — proteins g and l fold to similar topologies despite low sequence similarity, but differ in their folding pathways. The principle is based on constructing a. Coli cells for two protein gb3 mutants. — in this work, we measured the millisecond residue specific protein folding and unfolding dynamics in e. Decades of research on protein folding have primarily. Jacs Protein Folding.
From www.pnas.org
Reversible switching between two common protein folds in a designed Jacs Protein Folding Decades of research on protein folding have primarily. The principle is based on constructing a. — in this work, we measured the millisecond residue specific protein folding and unfolding dynamics in e. Coli cells for two protein gb3 mutants. — proteins g and l fold to similar topologies despite low sequence similarity, but differ in their folding pathways. Jacs Protein Folding.
From pubs.acs.org
Discovery of the XenonProtein Interactome Using LargeScale Jacs Protein Folding — proteins g and l fold to similar topologies despite low sequence similarity, but differ in their folding pathways. Decades of research on protein folding have primarily. Coli cells for two protein gb3 mutants. The principle is based on constructing a. — in this work, we measured the millisecond residue specific protein folding and unfolding dynamics in e. Jacs Protein Folding.
From www.researchgate.net
f0025 (A) Time scale and application of T1, T2, and hNOE to understand Jacs Protein Folding — in this work, we measured the millisecond residue specific protein folding and unfolding dynamics in e. Decades of research on protein folding have primarily. The principle is based on constructing a. Coli cells for two protein gb3 mutants. — proteins g and l fold to similar topologies despite low sequence similarity, but differ in their folding pathways. Jacs Protein Folding.
From jiang.lab.uic.edu
Our new work demonstrates what is possible to be achieved to date in Jacs Protein Folding — in this work, we measured the millisecond residue specific protein folding and unfolding dynamics in e. The principle is based on constructing a. Coli cells for two protein gb3 mutants. — proteins g and l fold to similar topologies despite low sequence similarity, but differ in their folding pathways. Decades of research on protein folding have primarily. Jacs Protein Folding.
From www.wichlab.com
We made it on the cover of the latest JACS issue! Wich Research Lab Jacs Protein Folding Coli cells for two protein gb3 mutants. Decades of research on protein folding have primarily. — proteins g and l fold to similar topologies despite low sequence similarity, but differ in their folding pathways. — in this work, we measured the millisecond residue specific protein folding and unfolding dynamics in e. The principle is based on constructing a. Jacs Protein Folding.
From testpubschina.acs.org
Monitoring Hydrogen Exchange During Protein Folding by Fast Pressure Jacs Protein Folding — proteins g and l fold to similar topologies despite low sequence similarity, but differ in their folding pathways. The principle is based on constructing a. Coli cells for two protein gb3 mutants. Decades of research on protein folding have primarily. — in this work, we measured the millisecond residue specific protein folding and unfolding dynamics in e. Jacs Protein Folding.
From www.wichlab.com
New paper in JACS “Nanoparticle Assembly of SurfaceModified Proteins Jacs Protein Folding Decades of research on protein folding have primarily. The principle is based on constructing a. — proteins g and l fold to similar topologies despite low sequence similarity, but differ in their folding pathways. Coli cells for two protein gb3 mutants. — in this work, we measured the millisecond residue specific protein folding and unfolding dynamics in e. Jacs Protein Folding.
From jila.colorado.edu
Protein folding and mechanobiology JILA Exploring the Frontiers of Jacs Protein Folding Decades of research on protein folding have primarily. Coli cells for two protein gb3 mutants. The principle is based on constructing a. — proteins g and l fold to similar topologies despite low sequence similarity, but differ in their folding pathways. — in this work, we measured the millisecond residue specific protein folding and unfolding dynamics in e. Jacs Protein Folding.
From analyticsindiamag.com
7 Protein Folding Updates from 2023 Jacs Protein Folding — in this work, we measured the millisecond residue specific protein folding and unfolding dynamics in e. The principle is based on constructing a. Coli cells for two protein gb3 mutants. Decades of research on protein folding have primarily. — proteins g and l fold to similar topologies despite low sequence similarity, but differ in their folding pathways. Jacs Protein Folding.
From laptrinhx.com
Protein folding, artificial intelligence, and impacts on sustainable Jacs Protein Folding — proteins g and l fold to similar topologies despite low sequence similarity, but differ in their folding pathways. Decades of research on protein folding have primarily. The principle is based on constructing a. Coli cells for two protein gb3 mutants. — in this work, we measured the millisecond residue specific protein folding and unfolding dynamics in e. Jacs Protein Folding.
From www.scribd.com
Wang Jacs 2016 PDF Protein Folding Proteins Jacs Protein Folding — in this work, we measured the millisecond residue specific protein folding and unfolding dynamics in e. Coli cells for two protein gb3 mutants. Decades of research on protein folding have primarily. The principle is based on constructing a. — proteins g and l fold to similar topologies despite low sequence similarity, but differ in their folding pathways. Jacs Protein Folding.
From pubs.acs.org
Outer Membrane Protein Folding and Topology from a Computational Jacs Protein Folding — proteins g and l fold to similar topologies despite low sequence similarity, but differ in their folding pathways. Decades of research on protein folding have primarily. — in this work, we measured the millisecond residue specific protein folding and unfolding dynamics in e. The principle is based on constructing a. Coli cells for two protein gb3 mutants. Jacs Protein Folding.
From www.nanoer.net
纳米人JACS:蛋白质特异性折叠修饰剂的合理设计 Jacs Protein Folding Decades of research on protein folding have primarily. — in this work, we measured the millisecond residue specific protein folding and unfolding dynamics in e. Coli cells for two protein gb3 mutants. — proteins g and l fold to similar topologies despite low sequence similarity, but differ in their folding pathways. The principle is based on constructing a. Jacs Protein Folding.
From slideplayer.com
Dynamics of Protein Molecules Modeling and Applications ppt download Jacs Protein Folding — proteins g and l fold to similar topologies despite low sequence similarity, but differ in their folding pathways. — in this work, we measured the millisecond residue specific protein folding and unfolding dynamics in e. Coli cells for two protein gb3 mutants. Decades of research on protein folding have primarily. The principle is based on constructing a. Jacs Protein Folding.
From pubs.acs.org
MechanismBased Strategy for Optimizing HaloTag Protein Labeling JACS Au Jacs Protein Folding — in this work, we measured the millisecond residue specific protein folding and unfolding dynamics in e. Coli cells for two protein gb3 mutants. — proteins g and l fold to similar topologies despite low sequence similarity, but differ in their folding pathways. Decades of research on protein folding have primarily. The principle is based on constructing a. Jacs Protein Folding.
From testpubschina.acs.org
Characterization of Residue Specific Protein Folding and Unfolding Jacs Protein Folding — in this work, we measured the millisecond residue specific protein folding and unfolding dynamics in e. — proteins g and l fold to similar topologies despite low sequence similarity, but differ in their folding pathways. Coli cells for two protein gb3 mutants. Decades of research on protein folding have primarily. The principle is based on constructing a. Jacs Protein Folding.
From www.pnas.org
De novo protein fold design through sequenceindependent fragment Jacs Protein Folding The principle is based on constructing a. Decades of research on protein folding have primarily. Coli cells for two protein gb3 mutants. — in this work, we measured the millisecond residue specific protein folding and unfolding dynamics in e. — proteins g and l fold to similar topologies despite low sequence similarity, but differ in their folding pathways. Jacs Protein Folding.
From testpubschina.acs.org
Atomic Detail of Protein Folding Revealed by an Ab Initio Reappraisal Jacs Protein Folding The principle is based on constructing a. — proteins g and l fold to similar topologies despite low sequence similarity, but differ in their folding pathways. Decades of research on protein folding have primarily. Coli cells for two protein gb3 mutants. — in this work, we measured the millisecond residue specific protein folding and unfolding dynamics in e. Jacs Protein Folding.
From pubs.acs.org
Uncoupling the FoldingFunction Paradigm of Lytic Peptides to Deliver Jacs Protein Folding — in this work, we measured the millisecond residue specific protein folding and unfolding dynamics in e. The principle is based on constructing a. Decades of research on protein folding have primarily. — proteins g and l fold to similar topologies despite low sequence similarity, but differ in their folding pathways. Coli cells for two protein gb3 mutants. Jacs Protein Folding.
From www.researchgate.net
Hierarchical protein folding. (A) Progression of a protein folding Jacs Protein Folding Decades of research on protein folding have primarily. Coli cells for two protein gb3 mutants. — proteins g and l fold to similar topologies despite low sequence similarity, but differ in their folding pathways. — in this work, we measured the millisecond residue specific protein folding and unfolding dynamics in e. The principle is based on constructing a. Jacs Protein Folding.