Primer Blast Ensembl . Maximum of 30 sequences (type in plain text, fasta or sequence id) or upload sequence file. Blast [1] and blat [2] are sequence similarity search tools that can be used for both dna and proteins. The exons view provides information about specific exons and introns (figure 16). Blat is the default tool in ensembl. Ensembl annotate genes, computes multiple alignments, predicts regulatory function and collects disease data. Nous voudrions effectuer une description ici mais le site que vous consultez ne nous en laisse pas la possibilité. It may be useful for selecting sequence to design primers. If you do not have a gene name, an id, or an accession number for your sequence of interest, ensembl provides an interface that allows you. The blast tool allows any sequence to be compared against any genomic assembly in ensembl.
from www.slideshare.net
Blat is the default tool in ensembl. If you do not have a gene name, an id, or an accession number for your sequence of interest, ensembl provides an interface that allows you. It may be useful for selecting sequence to design primers. Ensembl annotate genes, computes multiple alignments, predicts regulatory function and collects disease data. Nous voudrions effectuer une description ici mais le site que vous consultez ne nous en laisse pas la possibilité. Blast [1] and blat [2] are sequence similarity search tools that can be used for both dna and proteins. The exons view provides information about specific exons and introns (figure 16). The blast tool allows any sequence to be compared against any genomic assembly in ensembl. Maximum of 30 sequences (type in plain text, fasta or sequence id) or upload sequence file.
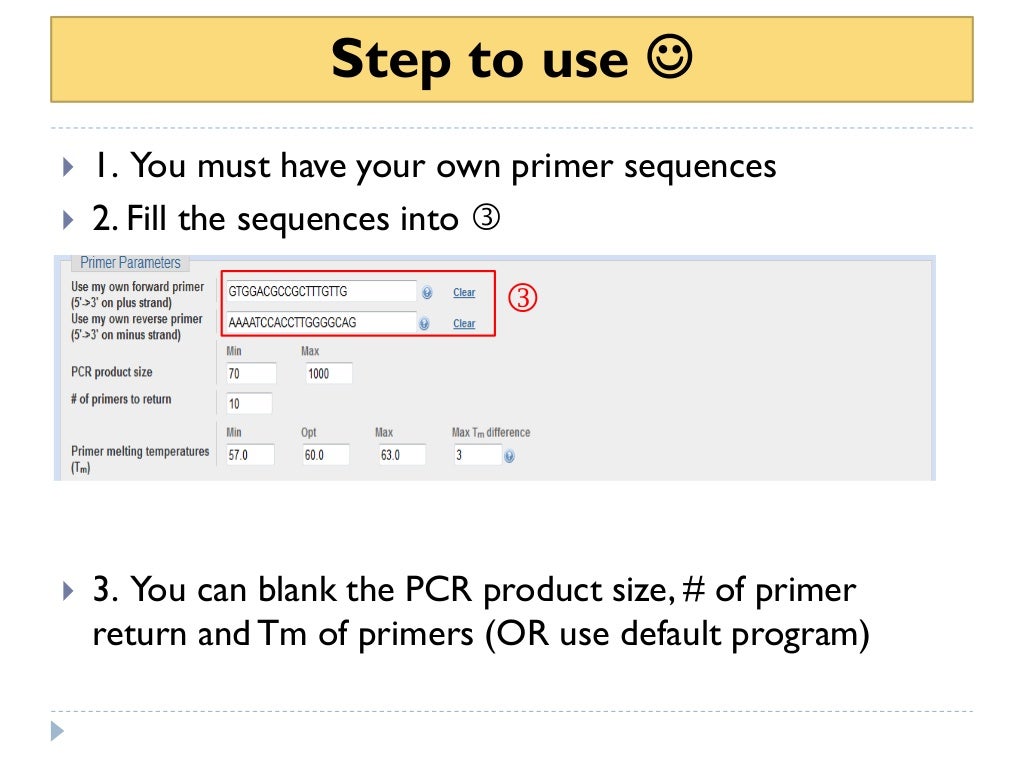
How to use primer blast for checking primer specificity
Primer Blast Ensembl If you do not have a gene name, an id, or an accession number for your sequence of interest, ensembl provides an interface that allows you. Blast [1] and blat [2] are sequence similarity search tools that can be used for both dna and proteins. Maximum of 30 sequences (type in plain text, fasta or sequence id) or upload sequence file. The blast tool allows any sequence to be compared against any genomic assembly in ensembl. The exons view provides information about specific exons and introns (figure 16). Nous voudrions effectuer une description ici mais le site que vous consultez ne nous en laisse pas la possibilité. Ensembl annotate genes, computes multiple alignments, predicts regulatory function and collects disease data. If you do not have a gene name, an id, or an accession number for your sequence of interest, ensembl provides an interface that allows you. Blat is the default tool in ensembl. It may be useful for selecting sequence to design primers.
From www.researchgate.net
Output of PrimerBLAST showing detailed primer reports Download Primer Blast Ensembl The exons view provides information about specific exons and introns (figure 16). Ensembl annotate genes, computes multiple alignments, predicts regulatory function and collects disease data. Blast [1] and blat [2] are sequence similarity search tools that can be used for both dna and proteins. It may be useful for selecting sequence to design primers. If you do not have a. Primer Blast Ensembl.
From www.researchgate.net
Primers/probes designed in silico using GenBank sequences (see Primer Blast Ensembl Blast [1] and blat [2] are sequence similarity search tools that can be used for both dna and proteins. The blast tool allows any sequence to be compared against any genomic assembly in ensembl. Maximum of 30 sequences (type in plain text, fasta or sequence id) or upload sequence file. If you do not have a gene name, an id,. Primer Blast Ensembl.
From www.researchgate.net
Primer sequences, Genbank accession number, Ensembl ID and amplicon Primer Blast Ensembl Maximum of 30 sequences (type in plain text, fasta or sequence id) or upload sequence file. The exons view provides information about specific exons and introns (figure 16). Ensembl annotate genes, computes multiple alignments, predicts regulatory function and collects disease data. Blast [1] and blat [2] are sequence similarity search tools that can be used for both dna and proteins.. Primer Blast Ensembl.
From www.youtube.com
How we can Design Primer? Primer Blast. Primer Specificity. forward and Primer Blast Ensembl The blast tool allows any sequence to be compared against any genomic assembly in ensembl. The exons view provides information about specific exons and introns (figure 16). Blast [1] and blat [2] are sequence similarity search tools that can be used for both dna and proteins. Blat is the default tool in ensembl. Maximum of 30 sequences (type in plain. Primer Blast Ensembl.
From www.nlm.nih.gov
Accessing PrimerBLAST Primer Blast Ensembl It may be useful for selecting sequence to design primers. Maximum of 30 sequences (type in plain text, fasta or sequence id) or upload sequence file. The exons view provides information about specific exons and introns (figure 16). Blat is the default tool in ensembl. The blast tool allows any sequence to be compared against any genomic assembly in ensembl.. Primer Blast Ensembl.
From www.nlm.nih.gov
Practice PrimerBLAST Examples Primer Blast Ensembl The blast tool allows any sequence to be compared against any genomic assembly in ensembl. Blast [1] and blat [2] are sequence similarity search tools that can be used for both dna and proteins. Nous voudrions effectuer une description ici mais le site que vous consultez ne nous en laisse pas la possibilité. Blat is the default tool in ensembl.. Primer Blast Ensembl.
From www.slideshare.net
How to use primer blast for checking primer specificity Primer Blast Ensembl Maximum of 30 sequences (type in plain text, fasta or sequence id) or upload sequence file. It may be useful for selecting sequence to design primers. If you do not have a gene name, an id, or an accession number for your sequence of interest, ensembl provides an interface that allows you. The blast tool allows any sequence to be. Primer Blast Ensembl.
From chanzuckerberg.zendesk.com
Prepare Primer BED Files CZ ID Help Center Primer Blast Ensembl The exons view provides information about specific exons and introns (figure 16). It may be useful for selecting sequence to design primers. If you do not have a gene name, an id, or an accession number for your sequence of interest, ensembl provides an interface that allows you. The blast tool allows any sequence to be compared against any genomic. Primer Blast Ensembl.
From www.nlm.nih.gov
Exercise 1 Identify the pathogen Primer Blast Ensembl The exons view provides information about specific exons and introns (figure 16). If you do not have a gene name, an id, or an accession number for your sequence of interest, ensembl provides an interface that allows you. It may be useful for selecting sequence to design primers. Maximum of 30 sequences (type in plain text, fasta or sequence id). Primer Blast Ensembl.
From ncbiinsights.ncbi.nlm.nih.gov
NCBI Insights ElectronicPCR (ePCR) is retiring, use PrimerBLAST Primer Blast Ensembl Blast [1] and blat [2] are sequence similarity search tools that can be used for both dna and proteins. Ensembl annotate genes, computes multiple alignments, predicts regulatory function and collects disease data. If you do not have a gene name, an id, or an accession number for your sequence of interest, ensembl provides an interface that allows you. It may. Primer Blast Ensembl.
From www.nlm.nih.gov
Accessing PrimerBLAST Primer Blast Ensembl If you do not have a gene name, an id, or an accession number for your sequence of interest, ensembl provides an interface that allows you. The blast tool allows any sequence to be compared against any genomic assembly in ensembl. Blat is the default tool in ensembl. Blast [1] and blat [2] are sequence similarity search tools that can. Primer Blast Ensembl.
From www.nlm.nih.gov
The PrimerBLAST interface Primer Blast Ensembl Maximum of 30 sequences (type in plain text, fasta or sequence id) or upload sequence file. The blast tool allows any sequence to be compared against any genomic assembly in ensembl. If you do not have a gene name, an id, or an accession number for your sequence of interest, ensembl provides an interface that allows you. Blat is the. Primer Blast Ensembl.
From zhuanlan.zhihu.com
如何使用 PrimerBlast 比对引物的特异性? (转载) 知乎 Primer Blast Ensembl If you do not have a gene name, an id, or an accession number for your sequence of interest, ensembl provides an interface that allows you. Blast [1] and blat [2] are sequence similarity search tools that can be used for both dna and proteins. Blat is the default tool in ensembl. Maximum of 30 sequences (type in plain text,. Primer Blast Ensembl.
From github.com
GitHub SimoesCostaLab/Primer_Tool A tool combining python, local Primer Blast Ensembl Blast [1] and blat [2] are sequence similarity search tools that can be used for both dna and proteins. It may be useful for selecting sequence to design primers. Blat is the default tool in ensembl. Nous voudrions effectuer une description ici mais le site que vous consultez ne nous en laisse pas la possibilité. The exons view provides information. Primer Blast Ensembl.
From www.nlm.nih.gov
Accessing PrimerBLAST Primer Blast Ensembl Ensembl annotate genes, computes multiple alignments, predicts regulatory function and collects disease data. Blast [1] and blat [2] are sequence similarity search tools that can be used for both dna and proteins. If you do not have a gene name, an id, or an accession number for your sequence of interest, ensembl provides an interface that allows you. Nous voudrions. Primer Blast Ensembl.
From www.nlm.nih.gov
Practice PrimerBLAST Examples Primer Blast Ensembl If you do not have a gene name, an id, or an accession number for your sequence of interest, ensembl provides an interface that allows you. Blast [1] and blat [2] are sequence similarity search tools that can be used for both dna and proteins. Blat is the default tool in ensembl. Maximum of 30 sequences (type in plain text,. Primer Blast Ensembl.
From toptipbio.com
NCBI Primer BLAST graphical view Top Tip Bio Primer Blast Ensembl The exons view provides information about specific exons and introns (figure 16). Maximum of 30 sequences (type in plain text, fasta or sequence id) or upload sequence file. Blat is the default tool in ensembl. Ensembl annotate genes, computes multiple alignments, predicts regulatory function and collects disease data. If you do not have a gene name, an id, or an. Primer Blast Ensembl.
From www.youtube.com
Primer BLAST Check your primers specificity using NCBI YouTube Primer Blast Ensembl The blast tool allows any sequence to be compared against any genomic assembly in ensembl. Ensembl annotate genes, computes multiple alignments, predicts regulatory function and collects disease data. Nous voudrions effectuer une description ici mais le site que vous consultez ne nous en laisse pas la possibilité. Blast [1] and blat [2] are sequence similarity search tools that can be. Primer Blast Ensembl.
From bio.libretexts.org
2.1.16.5 PrimerBLAST Biology LibreTexts Primer Blast Ensembl Ensembl annotate genes, computes multiple alignments, predicts regulatory function and collects disease data. Nous voudrions effectuer une description ici mais le site que vous consultez ne nous en laisse pas la possibilité. Maximum of 30 sequences (type in plain text, fasta or sequence id) or upload sequence file. The blast tool allows any sequence to be compared against any genomic. Primer Blast Ensembl.
From in.pinterest.com
Primer designing for real time PCR using NCBI Primer Blast Primer Primer Blast Ensembl Blast [1] and blat [2] are sequence similarity search tools that can be used for both dna and proteins. If you do not have a gene name, an id, or an accession number for your sequence of interest, ensembl provides an interface that allows you. The exons view provides information about specific exons and introns (figure 16). Nous voudrions effectuer. Primer Blast Ensembl.
From www.youtube.com
Primer Designing Using NCBI Primer Blast YouTube Primer Blast Ensembl Blat is the default tool in ensembl. If you do not have a gene name, an id, or an accession number for your sequence of interest, ensembl provides an interface that allows you. Ensembl annotate genes, computes multiple alignments, predicts regulatory function and collects disease data. The exons view provides information about specific exons and introns (figure 16). Maximum of. Primer Blast Ensembl.
From www.slideshare.net
How to use primer blast for checking primer specificity Primer Blast Ensembl Ensembl annotate genes, computes multiple alignments, predicts regulatory function and collects disease data. Maximum of 30 sequences (type in plain text, fasta or sequence id) or upload sequence file. Blast [1] and blat [2] are sequence similarity search tools that can be used for both dna and proteins. Blat is the default tool in ensembl. If you do not have. Primer Blast Ensembl.
From studylib.net
Introduction to NCBI & Ensembl tools including BLAST and database searching Primer Blast Ensembl Blat is the default tool in ensembl. Maximum of 30 sequences (type in plain text, fasta or sequence id) or upload sequence file. It may be useful for selecting sequence to design primers. Nous voudrions effectuer une description ici mais le site que vous consultez ne nous en laisse pas la possibilité. Ensembl annotate genes, computes multiple alignments, predicts regulatory. Primer Blast Ensembl.
From www.researchgate.net
List of PCR primers designed using NCBI/PrimerBLAST program Download Primer Blast Ensembl Ensembl annotate genes, computes multiple alignments, predicts regulatory function and collects disease data. Blast [1] and blat [2] are sequence similarity search tools that can be used for both dna and proteins. Nous voudrions effectuer une description ici mais le site que vous consultez ne nous en laisse pas la possibilité. It may be useful for selecting sequence to design. Primer Blast Ensembl.
From www.ncbi.nlm.nih.gov
Figure 1, PrimerBLAST results for UNG transcript variant 2 NCBI News Primer Blast Ensembl Maximum of 30 sequences (type in plain text, fasta or sequence id) or upload sequence file. Ensembl annotate genes, computes multiple alignments, predicts regulatory function and collects disease data. If you do not have a gene name, an id, or an accession number for your sequence of interest, ensembl provides an interface that allows you. Blat is the default tool. Primer Blast Ensembl.
From www.youtube.com
Primer Designing Using NCBI Primer Blast Tutorial 3a YouTube Primer Blast Ensembl Nous voudrions effectuer une description ici mais le site que vous consultez ne nous en laisse pas la possibilité. Blast [1] and blat [2] are sequence similarity search tools that can be used for both dna and proteins. Blat is the default tool in ensembl. The blast tool allows any sequence to be compared against any genomic assembly in ensembl.. Primer Blast Ensembl.
From www.slideshare.net
How to use primer blast for checking primer specificity Primer Blast Ensembl Blast [1] and blat [2] are sequence similarity search tools that can be used for both dna and proteins. It may be useful for selecting sequence to design primers. The exons view provides information about specific exons and introns (figure 16). The blast tool allows any sequence to be compared against any genomic assembly in ensembl. Ensembl annotate genes, computes. Primer Blast Ensembl.
From www.slideshare.net
How to use primer blast for checking primer specificity Primer Blast Ensembl It may be useful for selecting sequence to design primers. Nous voudrions effectuer une description ici mais le site que vous consultez ne nous en laisse pas la possibilité. Blast [1] and blat [2] are sequence similarity search tools that can be used for both dna and proteins. The exons view provides information about specific exons and introns (figure 16).. Primer Blast Ensembl.
From www.congress-intercultural.eu
Using Primer BLAST NCBI Video Vault NCBI Bookshelf, 42 OFF Primer Blast Ensembl The exons view provides information about specific exons and introns (figure 16). If you do not have a gene name, an id, or an accession number for your sequence of interest, ensembl provides an interface that allows you. Nous voudrions effectuer une description ici mais le site que vous consultez ne nous en laisse pas la possibilité. Maximum of 30. Primer Blast Ensembl.
From www.youtube.com
How to design primers using Primer Blast (NCBI) and check their Primer Blast Ensembl Ensembl annotate genes, computes multiple alignments, predicts regulatory function and collects disease data. Maximum of 30 sequences (type in plain text, fasta or sequence id) or upload sequence file. It may be useful for selecting sequence to design primers. If you do not have a gene name, an id, or an accession number for your sequence of interest, ensembl provides. Primer Blast Ensembl.
From www.youtube.com
How To Create RealTime PCR Primers Using PrimerBLAST YouTube Primer Blast Ensembl It may be useful for selecting sequence to design primers. Ensembl annotate genes, computes multiple alignments, predicts regulatory function and collects disease data. Maximum of 30 sequences (type in plain text, fasta or sequence id) or upload sequence file. The blast tool allows any sequence to be compared against any genomic assembly in ensembl. Blat is the default tool in. Primer Blast Ensembl.
From www.youtube.com
PrimerBLASTでPCR実験で使うプライマーを設計してみよう!(2020年7月版) YouTube Primer Blast Ensembl Blast [1] and blat [2] are sequence similarity search tools that can be used for both dna and proteins. Ensembl annotate genes, computes multiple alignments, predicts regulatory function and collects disease data. Maximum of 30 sequences (type in plain text, fasta or sequence id) or upload sequence file. It may be useful for selecting sequence to design primers. If you. Primer Blast Ensembl.
From www.youtube.com
primer design using NCBI Primer BLAST YouTube Primer Blast Ensembl Nous voudrions effectuer une description ici mais le site que vous consultez ne nous en laisse pas la possibilité. Blast [1] and blat [2] are sequence similarity search tools that can be used for both dna and proteins. The blast tool allows any sequence to be compared against any genomic assembly in ensembl. The exons view provides information about specific. Primer Blast Ensembl.
From www.youtube.com
PCR Primer Designing using NCBI PrimerBLAST YouTube Primer Blast Ensembl The exons view provides information about specific exons and introns (figure 16). Maximum of 30 sequences (type in plain text, fasta or sequence id) or upload sequence file. Nous voudrions effectuer une description ici mais le site que vous consultez ne nous en laisse pas la possibilité. Ensembl annotate genes, computes multiple alignments, predicts regulatory function and collects disease data.. Primer Blast Ensembl.
From ncbiinsights.ncbi.nlm.nih.gov
Using NCBI’s PrimerBLAST to design and analyze PCR primers NCBI Insights Primer Blast Ensembl Maximum of 30 sequences (type in plain text, fasta or sequence id) or upload sequence file. Nous voudrions effectuer une description ici mais le site que vous consultez ne nous en laisse pas la possibilité. It may be useful for selecting sequence to design primers. Ensembl annotate genes, computes multiple alignments, predicts regulatory function and collects disease data. If you. Primer Blast Ensembl.