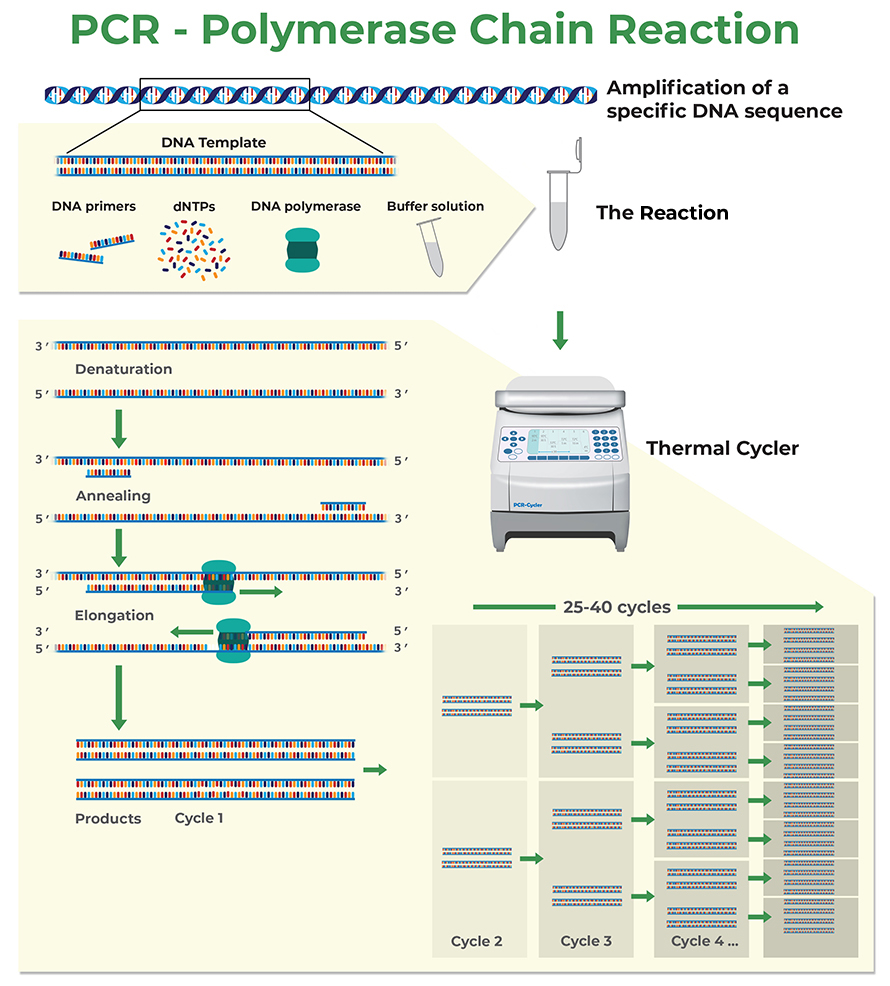
How To Use Primers In Pcr . Optimal primer sequences and appropriate primer concentrations are essential for maximal specificity and efficiency in pcr. They are usually 18 to 25. A standard pcr uses two primers, often called the “forward” and “reverse” primers. The forward and reverse primers are oriented on opposite. Here are some guidelines for designing your pcr primers: Enter the target sequence in fasta format or an accession number of an ncbi nucleotide sequence in the pcr template section of the form. Aim for the gc content to be between 40 and 60% with the 3’ of a primer ending in g or c to promote binding. Primers are short stretches of dna that target unique sequences of a dna molecule and help identify a unique part of a genome or a gene to be copied further. For pcr primer design, idt recommends that you aim for pcr primers between 18 and 30 bases; However, the most important considerations for.
from www.zymoresearch.com
However, the most important considerations for. For pcr primer design, idt recommends that you aim for pcr primers between 18 and 30 bases; Here are some guidelines for designing your pcr primers: Optimal primer sequences and appropriate primer concentrations are essential for maximal specificity and efficiency in pcr. They are usually 18 to 25. A standard pcr uses two primers, often called the “forward” and “reverse” primers. Primers are short stretches of dna that target unique sequences of a dna molecule and help identify a unique part of a genome or a gene to be copied further. Aim for the gc content to be between 40 and 60% with the 3’ of a primer ending in g or c to promote binding. Enter the target sequence in fasta format or an accession number of an ncbi nucleotide sequence in the pcr template section of the form. The forward and reverse primers are oriented on opposite.
How to Design Primers ZYMO RESEARCH
How To Use Primers In Pcr The forward and reverse primers are oriented on opposite. Optimal primer sequences and appropriate primer concentrations are essential for maximal specificity and efficiency in pcr. For pcr primer design, idt recommends that you aim for pcr primers between 18 and 30 bases; Primers are short stretches of dna that target unique sequences of a dna molecule and help identify a unique part of a genome or a gene to be copied further. Enter the target sequence in fasta format or an accession number of an ncbi nucleotide sequence in the pcr template section of the form. Here are some guidelines for designing your pcr primers: However, the most important considerations for. Aim for the gc content to be between 40 and 60% with the 3’ of a primer ending in g or c to promote binding. They are usually 18 to 25. A standard pcr uses two primers, often called the “forward” and “reverse” primers. The forward and reverse primers are oriented on opposite.
From www.bosterbio.com
Polymerase Chain Reaction (PCR) Key Principles How To Use Primers In Pcr Aim for the gc content to be between 40 and 60% with the 3’ of a primer ending in g or c to promote binding. The forward and reverse primers are oriented on opposite. Optimal primer sequences and appropriate primer concentrations are essential for maximal specificity and efficiency in pcr. They are usually 18 to 25. Primers are short stretches. How To Use Primers In Pcr.
From loemmmosi.blob.core.windows.net
Primer Dna Pcr at Linda South blog How To Use Primers In Pcr Primers are short stretches of dna that target unique sequences of a dna molecule and help identify a unique part of a genome or a gene to be copied further. Here are some guidelines for designing your pcr primers: Optimal primer sequences and appropriate primer concentrations are essential for maximal specificity and efficiency in pcr. For pcr primer design, idt. How To Use Primers In Pcr.
From ar.inspiredpencil.com
Primers Pcr How To Use Primers In Pcr Enter the target sequence in fasta format or an accession number of an ncbi nucleotide sequence in the pcr template section of the form. They are usually 18 to 25. Aim for the gc content to be between 40 and 60% with the 3’ of a primer ending in g or c to promote binding. However, the most important considerations. How To Use Primers In Pcr.
From www.researchgate.net
PCR using sequencespecific primers (PCRSSP) results for lowand How To Use Primers In Pcr The forward and reverse primers are oriented on opposite. They are usually 18 to 25. For pcr primer design, idt recommends that you aim for pcr primers between 18 and 30 bases; Aim for the gc content to be between 40 and 60% with the 3’ of a primer ending in g or c to promote binding. A standard pcr. How To Use Primers In Pcr.
From the-dna-universe.com
Primer design guide 5 tips for best PCR results How To Use Primers In Pcr Here are some guidelines for designing your pcr primers: Primers are short stretches of dna that target unique sequences of a dna molecule and help identify a unique part of a genome or a gene to be copied further. Optimal primer sequences and appropriate primer concentrations are essential for maximal specificity and efficiency in pcr. A standard pcr uses two. How To Use Primers In Pcr.
From www.addgene.org
Addgene What is Polymerase Chain Reaction (PCR) How To Use Primers In Pcr The forward and reverse primers are oriented on opposite. Primers are short stretches of dna that target unique sequences of a dna molecule and help identify a unique part of a genome or a gene to be copied further. Enter the target sequence in fasta format or an accession number of an ncbi nucleotide sequence in the pcr template section. How To Use Primers In Pcr.
From s-stembrendaarvizu.blogspot.com
SSTEM Scholar Brenda Arvizu's Blog PCR Primers How To Use Primers In Pcr For pcr primer design, idt recommends that you aim for pcr primers between 18 and 30 bases; However, the most important considerations for. Here are some guidelines for designing your pcr primers: Optimal primer sequences and appropriate primer concentrations are essential for maximal specificity and efficiency in pcr. Aim for the gc content to be between 40 and 60% with. How To Use Primers In Pcr.
From geneticeducation.co.in
How to Design PCR Primers How To Use Primers In Pcr Enter the target sequence in fasta format or an accession number of an ncbi nucleotide sequence in the pcr template section of the form. The forward and reverse primers are oriented on opposite. They are usually 18 to 25. Aim for the gc content to be between 40 and 60% with the 3’ of a primer ending in g or. How To Use Primers In Pcr.
From www.zymoresearch.com
How to Design Primers ZYMO RESEARCH How To Use Primers In Pcr Enter the target sequence in fasta format or an accession number of an ncbi nucleotide sequence in the pcr template section of the form. Primers are short stretches of dna that target unique sequences of a dna molecule and help identify a unique part of a genome or a gene to be copied further. Aim for the gc content to. How To Use Primers In Pcr.
From www.researchgate.net
PCR amplification using an ISSR primer Download Scientific Diagram How To Use Primers In Pcr Here are some guidelines for designing your pcr primers: Primers are short stretches of dna that target unique sequences of a dna molecule and help identify a unique part of a genome or a gene to be copied further. However, the most important considerations for. A standard pcr uses two primers, often called the “forward” and “reverse” primers. They are. How To Use Primers In Pcr.
From www.researchgate.net
How mutation at PCR primer binding sites gives rise to different PCR How To Use Primers In Pcr The forward and reverse primers are oriented on opposite. For pcr primer design, idt recommends that you aim for pcr primers between 18 and 30 bases; Optimal primer sequences and appropriate primer concentrations are essential for maximal specificity and efficiency in pcr. Primers are short stretches of dna that target unique sequences of a dna molecule and help identify a. How To Use Primers In Pcr.
From robersonquidents.blogspot.com
Pcr PCR primers...a primer! Virology Down Under Roberson Quidents How To Use Primers In Pcr They are usually 18 to 25. Aim for the gc content to be between 40 and 60% with the 3’ of a primer ending in g or c to promote binding. However, the most important considerations for. A standard pcr uses two primers, often called the “forward” and “reverse” primers. Primers are short stretches of dna that target unique sequences. How To Use Primers In Pcr.
From www.youtube.com
How To Create RealTime PCR Primers Using PrimerBLAST YouTube How To Use Primers In Pcr Optimal primer sequences and appropriate primer concentrations are essential for maximal specificity and efficiency in pcr. Primers are short stretches of dna that target unique sequences of a dna molecule and help identify a unique part of a genome or a gene to be copied further. Here are some guidelines for designing your pcr primers: A standard pcr uses two. How To Use Primers In Pcr.
From joisywxcy.blob.core.windows.net
Use Of Primer In Pcr at Evelyn Brown blog How To Use Primers In Pcr For pcr primer design, idt recommends that you aim for pcr primers between 18 and 30 bases; However, the most important considerations for. A standard pcr uses two primers, often called the “forward” and “reverse” primers. Here are some guidelines for designing your pcr primers: They are usually 18 to 25. The forward and reverse primers are oriented on opposite.. How To Use Primers In Pcr.
From www.minipcr.com
How to order PCR primers miniPCR bio How To Use Primers In Pcr For pcr primer design, idt recommends that you aim for pcr primers between 18 and 30 bases; The forward and reverse primers are oriented on opposite. They are usually 18 to 25. A standard pcr uses two primers, often called the “forward” and “reverse” primers. Optimal primer sequences and appropriate primer concentrations are essential for maximal specificity and efficiency in. How To Use Primers In Pcr.
From askbd.blogspot.com
Ask Polymerase chain reaction (PCR) How To Use Primers In Pcr For pcr primer design, idt recommends that you aim for pcr primers between 18 and 30 bases; The forward and reverse primers are oriented on opposite. Aim for the gc content to be between 40 and 60% with the 3’ of a primer ending in g or c to promote binding. A standard pcr uses two primers, often called the. How To Use Primers In Pcr.
From blog.addgene.org
Polymerase Chain Reaction Overview and Applications How To Use Primers In Pcr They are usually 18 to 25. The forward and reverse primers are oriented on opposite. A standard pcr uses two primers, often called the “forward” and “reverse” primers. Here are some guidelines for designing your pcr primers: Optimal primer sequences and appropriate primer concentrations are essential for maximal specificity and efficiency in pcr. For pcr primer design, idt recommends that. How To Use Primers In Pcr.
From www.researchgate.net
DNA sequence of the primers used for PCR amplification of Big domains How To Use Primers In Pcr Aim for the gc content to be between 40 and 60% with the 3’ of a primer ending in g or c to promote binding. For pcr primer design, idt recommends that you aim for pcr primers between 18 and 30 bases; Primers are short stretches of dna that target unique sequences of a dna molecule and help identify a. How To Use Primers In Pcr.
From www.researchgate.net
How to identify the efficiency of the primers used for PCR analysis How To Use Primers In Pcr The forward and reverse primers are oriented on opposite. Optimal primer sequences and appropriate primer concentrations are essential for maximal specificity and efficiency in pcr. Here are some guidelines for designing your pcr primers: They are usually 18 to 25. A standard pcr uses two primers, often called the “forward” and “reverse” primers. For pcr primer design, idt recommends that. How To Use Primers In Pcr.
From www.biobasic.com
Everything You Need to Know about PCR Primer Synthesis How To Use Primers In Pcr The forward and reverse primers are oriented on opposite. Enter the target sequence in fasta format or an accession number of an ncbi nucleotide sequence in the pcr template section of the form. A standard pcr uses two primers, often called the “forward” and “reverse” primers. For pcr primer design, idt recommends that you aim for pcr primers between 18. How To Use Primers In Pcr.
From joisywxcy.blob.core.windows.net
Use Of Primer In Pcr at Evelyn Brown blog How To Use Primers In Pcr Here are some guidelines for designing your pcr primers: Primers are short stretches of dna that target unique sequences of a dna molecule and help identify a unique part of a genome or a gene to be copied further. They are usually 18 to 25. Enter the target sequence in fasta format or an accession number of an ncbi nucleotide. How To Use Primers In Pcr.
From www.researchgate.net
Schematic diagram of nested PCR The first pair of primers amplifies How To Use Primers In Pcr Primers are short stretches of dna that target unique sequences of a dna molecule and help identify a unique part of a genome or a gene to be copied further. A standard pcr uses two primers, often called the “forward” and “reverse” primers. Enter the target sequence in fasta format or an accession number of an ncbi nucleotide sequence in. How To Use Primers In Pcr.
From www.youtube.com
How to choose Primers for PCR YouTube How To Use Primers In Pcr However, the most important considerations for. Optimal primer sequences and appropriate primer concentrations are essential for maximal specificity and efficiency in pcr. For pcr primer design, idt recommends that you aim for pcr primers between 18 and 30 bases; They are usually 18 to 25. The forward and reverse primers are oriented on opposite. Primers are short stretches of dna. How To Use Primers In Pcr.
From geneticeducation.co.in
How to Design PCR Primers How To Use Primers In Pcr Here are some guidelines for designing your pcr primers: For pcr primer design, idt recommends that you aim for pcr primers between 18 and 30 bases; The forward and reverse primers are oriented on opposite. Aim for the gc content to be between 40 and 60% with the 3’ of a primer ending in g or c to promote binding.. How To Use Primers In Pcr.
From geneticeducation.co.in
How to Design PCR Primers How To Use Primers In Pcr Primers are short stretches of dna that target unique sequences of a dna molecule and help identify a unique part of a genome or a gene to be copied further. A standard pcr uses two primers, often called the “forward” and “reverse” primers. They are usually 18 to 25. Optimal primer sequences and appropriate primer concentrations are essential for maximal. How To Use Primers In Pcr.
From www.goldbio.com
How to choose a PCR Probe GoldBio How To Use Primers In Pcr Optimal primer sequences and appropriate primer concentrations are essential for maximal specificity and efficiency in pcr. A standard pcr uses two primers, often called the “forward” and “reverse” primers. Enter the target sequence in fasta format or an accession number of an ncbi nucleotide sequence in the pcr template section of the form. For pcr primer design, idt recommends that. How To Use Primers In Pcr.
From www.onlinebiologynotes.com
Polymerase chain reaction (PCR) Principle, procedure or steps, types How To Use Primers In Pcr Aim for the gc content to be between 40 and 60% with the 3’ of a primer ending in g or c to promote binding. The forward and reverse primers are oriented on opposite. They are usually 18 to 25. However, the most important considerations for. Optimal primer sequences and appropriate primer concentrations are essential for maximal specificity and efficiency. How To Use Primers In Pcr.
From www.sigmaaldrich.com
PCR Applications How To Use Primers In Pcr Here are some guidelines for designing your pcr primers: A standard pcr uses two primers, often called the “forward” and “reverse” primers. Optimal primer sequences and appropriate primer concentrations are essential for maximal specificity and efficiency in pcr. Enter the target sequence in fasta format or an accession number of an ncbi nucleotide sequence in the pcr template section of. How To Use Primers In Pcr.
From www.pinterest.com
Tutorial 1 Designing basic PCR primers Primer, Tutorial, Basic concepts How To Use Primers In Pcr A standard pcr uses two primers, often called the “forward” and “reverse” primers. Primers are short stretches of dna that target unique sequences of a dna molecule and help identify a unique part of a genome or a gene to be copied further. Enter the target sequence in fasta format or an accession number of an ncbi nucleotide sequence in. How To Use Primers In Pcr.
From www.thermofisher.com
Traditional Cloning Basics Thermo Fisher Scientific DE How To Use Primers In Pcr However, the most important considerations for. They are usually 18 to 25. For pcr primer design, idt recommends that you aim for pcr primers between 18 and 30 bases; Primers are short stretches of dna that target unique sequences of a dna molecule and help identify a unique part of a genome or a gene to be copied further. A. How To Use Primers In Pcr.
From www.technologynetworks.com
An Introduction to PCR Technology Networks How To Use Primers In Pcr Optimal primer sequences and appropriate primer concentrations are essential for maximal specificity and efficiency in pcr. For pcr primer design, idt recommends that you aim for pcr primers between 18 and 30 bases; Here are some guidelines for designing your pcr primers: A standard pcr uses two primers, often called the “forward” and “reverse” primers. Enter the target sequence in. How To Use Primers In Pcr.
From www.zymoresearch.com
How to Design Primers ZYMO RESEARCH How To Use Primers In Pcr For pcr primer design, idt recommends that you aim for pcr primers between 18 and 30 bases; Aim for the gc content to be between 40 and 60% with the 3’ of a primer ending in g or c to promote binding. Here are some guidelines for designing your pcr primers: The forward and reverse primers are oriented on opposite.. How To Use Primers In Pcr.
From www.researchgate.net
5 Principle of the PCR in which small specific DNA sequences (primers How To Use Primers In Pcr Primers are short stretches of dna that target unique sequences of a dna molecule and help identify a unique part of a genome or a gene to be copied further. Enter the target sequence in fasta format or an accession number of an ncbi nucleotide sequence in the pcr template section of the form. Here are some guidelines for designing. How To Use Primers In Pcr.
From www.researchgate.net
Schematic of sgRNA synthesis and threeprimer PCR strategies for How To Use Primers In Pcr Enter the target sequence in fasta format or an accession number of an ncbi nucleotide sequence in the pcr template section of the form. A standard pcr uses two primers, often called the “forward” and “reverse” primers. Aim for the gc content to be between 40 and 60% with the 3’ of a primer ending in g or c to. How To Use Primers In Pcr.
From www.researchgate.net
Schematic representation of overlap extension PCR. Two DNA fragments How To Use Primers In Pcr Here are some guidelines for designing your pcr primers: However, the most important considerations for. A standard pcr uses two primers, often called the “forward” and “reverse” primers. Optimal primer sequences and appropriate primer concentrations are essential for maximal specificity and efficiency in pcr. The forward and reverse primers are oriented on opposite. Aim for the gc content to be. How To Use Primers In Pcr.