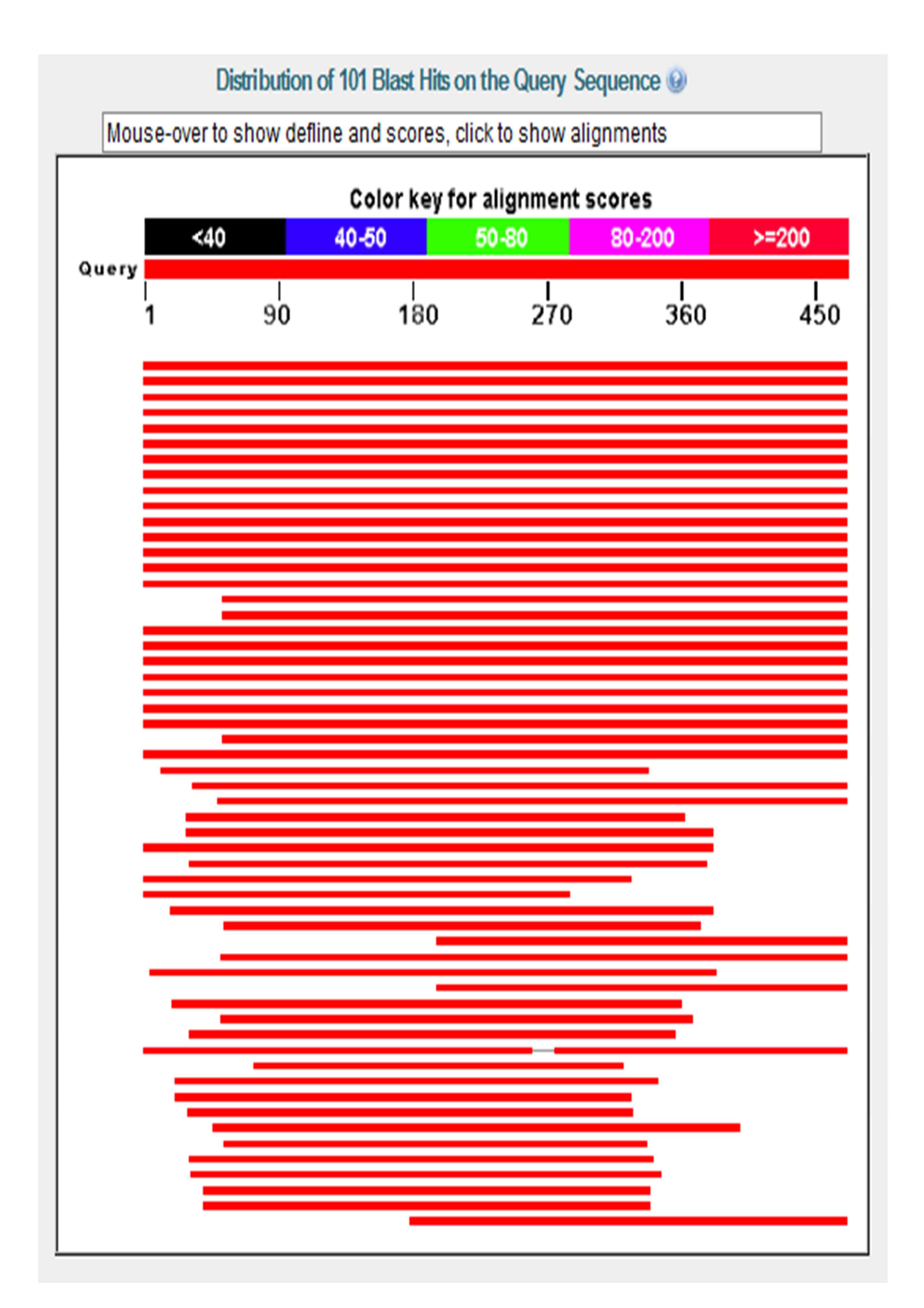
Protein Blast Embl . Blastp simply compares a protein query to a protein database. You can use smart in two different modes: Access the protein blast (basic local alignment search tool) tool below. Normal or genomic.the main difference is in the underlying protein database used. Blast stands for basic local alignment search tool.the emphasis of this tool is to find regions of sequence similarity, which will yield functional.
from www.ucl.ac.uk
Blastp simply compares a protein query to a protein database. Blast stands for basic local alignment search tool.the emphasis of this tool is to find regions of sequence similarity, which will yield functional. You can use smart in two different modes: Normal or genomic.the main difference is in the underlying protein database used. Access the protein blast (basic local alignment search tool) tool below.
BLAST
Protein Blast Embl You can use smart in two different modes: Blastp simply compares a protein query to a protein database. Normal or genomic.the main difference is in the underlying protein database used. Blast stands for basic local alignment search tool.the emphasis of this tool is to find regions of sequence similarity, which will yield functional. You can use smart in two different modes: Access the protein blast (basic local alignment search tool) tool below.
From www.frontiersin.org
Frontiers The Pepper MAP Kinase CaAIMK1 Positively Regulates ABA and Protein Blast Embl Access the protein blast (basic local alignment search tool) tool below. Blastp simply compares a protein query to a protein database. Normal or genomic.the main difference is in the underlying protein database used. Blast stands for basic local alignment search tool.the emphasis of this tool is to find regions of sequence similarity, which will yield functional. You can use smart. Protein Blast Embl.
From www.craiyon.com
Visualization of protein binding to dna and chromatin on Craiyon Protein Blast Embl Normal or genomic.the main difference is in the underlying protein database used. Access the protein blast (basic local alignment search tool) tool below. Blast stands for basic local alignment search tool.the emphasis of this tool is to find regions of sequence similarity, which will yield functional. Blastp simply compares a protein query to a protein database. You can use smart. Protein Blast Embl.
From abacus.bates.edu
BLASTP Results Part 1 Protein Blast Embl Blastp simply compares a protein query to a protein database. Normal or genomic.the main difference is in the underlying protein database used. Access the protein blast (basic local alignment search tool) tool below. Blast stands for basic local alignment search tool.the emphasis of this tool is to find regions of sequence similarity, which will yield functional. You can use smart. Protein Blast Embl.
From www.ncbi.nlm.nih.gov
NCBI Conserved Domain Database (CDD) Help Protein Blast Embl Normal or genomic.the main difference is in the underlying protein database used. Access the protein blast (basic local alignment search tool) tool below. You can use smart in two different modes: Blast stands for basic local alignment search tool.the emphasis of this tool is to find regions of sequence similarity, which will yield functional. Blastp simply compares a protein query. Protein Blast Embl.
From www.figma.com
Figma Protein Blast Embl You can use smart in two different modes: Blastp simply compares a protein query to a protein database. Normal or genomic.the main difference is in the underlying protein database used. Blast stands for basic local alignment search tool.the emphasis of this tool is to find regions of sequence similarity, which will yield functional. Access the protein blast (basic local alignment. Protein Blast Embl.
From cgp.iiarjournals.org
Analysis of ProteinProtein Interactions Identifies NECTIN2 as a Target Protein Blast Embl Access the protein blast (basic local alignment search tool) tool below. Normal or genomic.the main difference is in the underlying protein database used. You can use smart in two different modes: Blastp simply compares a protein query to a protein database. Blast stands for basic local alignment search tool.the emphasis of this tool is to find regions of sequence similarity,. Protein Blast Embl.
From www.researchgate.net
An example workflow from NCBI BLAST+ to Clustal Omega and construction Protein Blast Embl You can use smart in two different modes: Blast stands for basic local alignment search tool.the emphasis of this tool is to find regions of sequence similarity, which will yield functional. Blastp simply compares a protein query to a protein database. Normal or genomic.the main difference is in the underlying protein database used. Access the protein blast (basic local alignment. Protein Blast Embl.
From www.iherb.com
RYSE, Loaded Protein, Chocolate Cookie Blast, 2.3 lbs (1,056 g) Protein Blast Embl You can use smart in two different modes: Access the protein blast (basic local alignment search tool) tool below. Blast stands for basic local alignment search tool.the emphasis of this tool is to find regions of sequence similarity, which will yield functional. Blastp simply compares a protein query to a protein database. Normal or genomic.the main difference is in the. Protein Blast Embl.
From www.youtube.com
BLAST Protein YouTube Protein Blast Embl You can use smart in two different modes: Normal or genomic.the main difference is in the underlying protein database used. Blastp simply compares a protein query to a protein database. Access the protein blast (basic local alignment search tool) tool below. Blast stands for basic local alignment search tool.the emphasis of this tool is to find regions of sequence similarity,. Protein Blast Embl.
From kimbio.info
Protein blast における ‘Positives’ はどんな意味か? Kim Biology & Informatics Protein Blast Embl Access the protein blast (basic local alignment search tool) tool below. Normal or genomic.the main difference is in the underlying protein database used. Blastp simply compares a protein query to a protein database. Blast stands for basic local alignment search tool.the emphasis of this tool is to find regions of sequence similarity, which will yield functional. You can use smart. Protein Blast Embl.
From www.myprotein.com
Vimto Clear Whey Isolate Whey Protein Drink Myprotein Protein Blast Embl You can use smart in two different modes: Normal or genomic.the main difference is in the underlying protein database used. Blast stands for basic local alignment search tool.the emphasis of this tool is to find regions of sequence similarity, which will yield functional. Blastp simply compares a protein query to a protein database. Access the protein blast (basic local alignment. Protein Blast Embl.
From www.psb.ugent.be
PTM Viewer Protein Blast Embl Blastp simply compares a protein query to a protein database. Access the protein blast (basic local alignment search tool) tool below. You can use smart in two different modes: Normal or genomic.the main difference is in the underlying protein database used. Blast stands for basic local alignment search tool.the emphasis of this tool is to find regions of sequence similarity,. Protein Blast Embl.
From phys.org
Pro1 protein malfunction allows rice blast fungus to thrive, new study Protein Blast Embl Blast stands for basic local alignment search tool.the emphasis of this tool is to find regions of sequence similarity, which will yield functional. Normal or genomic.the main difference is in the underlying protein database used. Access the protein blast (basic local alignment search tool) tool below. Blastp simply compares a protein query to a protein database. You can use smart. Protein Blast Embl.
From pediaa.com
Difference Between BLAST and FASTA Definition, Features, Uses Protein Blast Embl You can use smart in two different modes: Blast stands for basic local alignment search tool.the emphasis of this tool is to find regions of sequence similarity, which will yield functional. Normal or genomic.the main difference is in the underlying protein database used. Blastp simply compares a protein query to a protein database. Access the protein blast (basic local alignment. Protein Blast Embl.
From www.researchgate.net
NCBI Protein BLAST Taxonomy OmpC321340 Download Scientific Diagram Protein Blast Embl Normal or genomic.the main difference is in the underlying protein database used. Access the protein blast (basic local alignment search tool) tool below. Blast stands for basic local alignment search tool.the emphasis of this tool is to find regions of sequence similarity, which will yield functional. Blastp simply compares a protein query to a protein database. You can use smart. Protein Blast Embl.
From www.trendradars.com
Bigg Boss 16 Week 1 Nomination Shiv Thakre, Sajid Khan,Gori Lagori Protein Blast Embl Blast stands for basic local alignment search tool.the emphasis of this tool is to find regions of sequence similarity, which will yield functional. Access the protein blast (basic local alignment search tool) tool below. Normal or genomic.the main difference is in the underlying protein database used. You can use smart in two different modes: Blastp simply compares a protein query. Protein Blast Embl.
From typeset.io
(Open Access) A dotmatrix program with dynamic threshold control Protein Blast Embl Blastp simply compares a protein query to a protein database. Access the protein blast (basic local alignment search tool) tool below. Blast stands for basic local alignment search tool.the emphasis of this tool is to find regions of sequence similarity, which will yield functional. You can use smart in two different modes: Normal or genomic.the main difference is in the. Protein Blast Embl.
From tcdb.org
TCBLAST Tutorial Protein Blast Embl Blast stands for basic local alignment search tool.the emphasis of this tool is to find regions of sequence similarity, which will yield functional. Blastp simply compares a protein query to a protein database. Access the protein blast (basic local alignment search tool) tool below. Normal or genomic.the main difference is in the underlying protein database used. You can use smart. Protein Blast Embl.
From sgbc.github.io
Blast Online Bioinformatics course Protein Blast Embl Blastp simply compares a protein query to a protein database. Blast stands for basic local alignment search tool.the emphasis of this tool is to find regions of sequence similarity, which will yield functional. Access the protein blast (basic local alignment search tool) tool below. You can use smart in two different modes: Normal or genomic.the main difference is in the. Protein Blast Embl.
From www.etprotein.com
Banana Blast Ultimate Protein for Explosive WorkoutsETprotein Protein Blast Embl You can use smart in two different modes: Blastp simply compares a protein query to a protein database. Normal or genomic.the main difference is in the underlying protein database used. Blast stands for basic local alignment search tool.the emphasis of this tool is to find regions of sequence similarity, which will yield functional. Access the protein blast (basic local alignment. Protein Blast Embl.
From www.explore.biobus.org
BLAST some proteins! BioBus STEAM Challenges Protein Blast Embl You can use smart in two different modes: Blast stands for basic local alignment search tool.the emphasis of this tool is to find regions of sequence similarity, which will yield functional. Access the protein blast (basic local alignment search tool) tool below. Blastp simply compares a protein query to a protein database. Normal or genomic.the main difference is in the. Protein Blast Embl.
From www.youtube.com
Translated Nucleotide to Protein BLAST BLASTX YouTube Protein Blast Embl Access the protein blast (basic local alignment search tool) tool below. You can use smart in two different modes: Blastp simply compares a protein query to a protein database. Blast stands for basic local alignment search tool.the emphasis of this tool is to find regions of sequence similarity, which will yield functional. Normal or genomic.the main difference is in the. Protein Blast Embl.
From teaching.ncl.ac.uk
BMN2000/CMB2000 Informatics Practical BLAST Protein Search Protein Blast Embl Access the protein blast (basic local alignment search tool) tool below. Blastp simply compares a protein query to a protein database. Blast stands for basic local alignment search tool.the emphasis of this tool is to find regions of sequence similarity, which will yield functional. You can use smart in two different modes: Normal or genomic.the main difference is in the. Protein Blast Embl.
From kimbio.info
Protein blast における ‘Positives’ はどんな意味か? Kim Biology & Informatics Protein Blast Embl Access the protein blast (basic local alignment search tool) tool below. Blastp simply compares a protein query to a protein database. Blast stands for basic local alignment search tool.the emphasis of this tool is to find regions of sequence similarity, which will yield functional. Normal or genomic.the main difference is in the underlying protein database used. You can use smart. Protein Blast Embl.
From www.youtube.com
Which Database to Choose for Protein BLAST run YouTube Protein Blast Embl Blast stands for basic local alignment search tool.the emphasis of this tool is to find regions of sequence similarity, which will yield functional. You can use smart in two different modes: Normal or genomic.the main difference is in the underlying protein database used. Access the protein blast (basic local alignment search tool) tool below. Blastp simply compares a protein query. Protein Blast Embl.
From www.researchgate.net
(PDF) Expression of nuclear pore protein POM121 in childhood acute Protein Blast Embl Blast stands for basic local alignment search tool.the emphasis of this tool is to find regions of sequence similarity, which will yield functional. Blastp simply compares a protein query to a protein database. Access the protein blast (basic local alignment search tool) tool below. You can use smart in two different modes: Normal or genomic.the main difference is in the. Protein Blast Embl.
From lacaleya.org
BLAST Resultsintroduktion till NCBI Bioinformatics ResourcesLearning Protein Blast Embl Blast stands for basic local alignment search tool.the emphasis of this tool is to find regions of sequence similarity, which will yield functional. Blastp simply compares a protein query to a protein database. You can use smart in two different modes: Access the protein blast (basic local alignment search tool) tool below. Normal or genomic.the main difference is in the. Protein Blast Embl.
From abc-supplements.com
FA Diamond Hydrolysed Whey Protein 2kg ABC Supplements Protein Blast Embl Normal or genomic.the main difference is in the underlying protein database used. Access the protein blast (basic local alignment search tool) tool below. Blastp simply compares a protein query to a protein database. You can use smart in two different modes: Blast stands for basic local alignment search tool.the emphasis of this tool is to find regions of sequence similarity,. Protein Blast Embl.
From studylib.net
Using BLAST to Analyze Fluorescent Proteins Protein Blast Embl Blastp simply compares a protein query to a protein database. Normal or genomic.the main difference is in the underlying protein database used. Access the protein blast (basic local alignment search tool) tool below. You can use smart in two different modes: Blast stands for basic local alignment search tool.the emphasis of this tool is to find regions of sequence similarity,. Protein Blast Embl.
From burlingtonchocolate.com
Protein Blast Burlington Chocolate Protein Blast Embl Access the protein blast (basic local alignment search tool) tool below. Blastp simply compares a protein query to a protein database. You can use smart in two different modes: Normal or genomic.the main difference is in the underlying protein database used. Blast stands for basic local alignment search tool.the emphasis of this tool is to find regions of sequence similarity,. Protein Blast Embl.
From www.stack3d.com
RESQ Stack3d Protein Blast Embl Blastp simply compares a protein query to a protein database. Blast stands for basic local alignment search tool.the emphasis of this tool is to find regions of sequence similarity, which will yield functional. Normal or genomic.the main difference is in the underlying protein database used. You can use smart in two different modes: Access the protein blast (basic local alignment. Protein Blast Embl.
From www.youtube.com
Which BLAST Algorithm to use for ProteinProtein BLAST YouTube Protein Blast Embl You can use smart in two different modes: Normal or genomic.the main difference is in the underlying protein database used. Blastp simply compares a protein query to a protein database. Blast stands for basic local alignment search tool.the emphasis of this tool is to find regions of sequence similarity, which will yield functional. Access the protein blast (basic local alignment. Protein Blast Embl.
From bookdown.org
Chapter 4 3 Predicting structure and function from sequence Protein Blast Embl Blastp simply compares a protein query to a protein database. Normal or genomic.the main difference is in the underlying protein database used. You can use smart in two different modes: Blast stands for basic local alignment search tool.the emphasis of this tool is to find regions of sequence similarity, which will yield functional. Access the protein blast (basic local alignment. Protein Blast Embl.
From www.ucl.ac.uk
BLAST Protein Blast Embl Blast stands for basic local alignment search tool.the emphasis of this tool is to find regions of sequence similarity, which will yield functional. Normal or genomic.the main difference is in the underlying protein database used. Access the protein blast (basic local alignment search tool) tool below. Blastp simply compares a protein query to a protein database. You can use smart. Protein Blast Embl.
From usuhs.libguides.com
BLASTP Protein Sequence Search Introduction to NCBI Bioinformatics Protein Blast Embl Blastp simply compares a protein query to a protein database. You can use smart in two different modes: Normal or genomic.the main difference is in the underlying protein database used. Access the protein blast (basic local alignment search tool) tool below. Blast stands for basic local alignment search tool.the emphasis of this tool is to find regions of sequence similarity,. Protein Blast Embl.