Eggnog Mapper Command Line . Emapper.py or just emapper) is a tool for fast functional annotation of novel sequences. It uses precomputed orthologous groups and. This function will read in the table and output a tidy table of one part of the table (eg., cog functional categories or kegg pathways). It uses precomputed orthologous groups and.
from blog.csdn.net
This function will read in the table and output a tidy table of one part of the table (eg., cog functional categories or kegg pathways). It uses precomputed orthologous groups and. Emapper.py or just emapper) is a tool for fast functional annotation of novel sequences. It uses precomputed orthologous groups and.
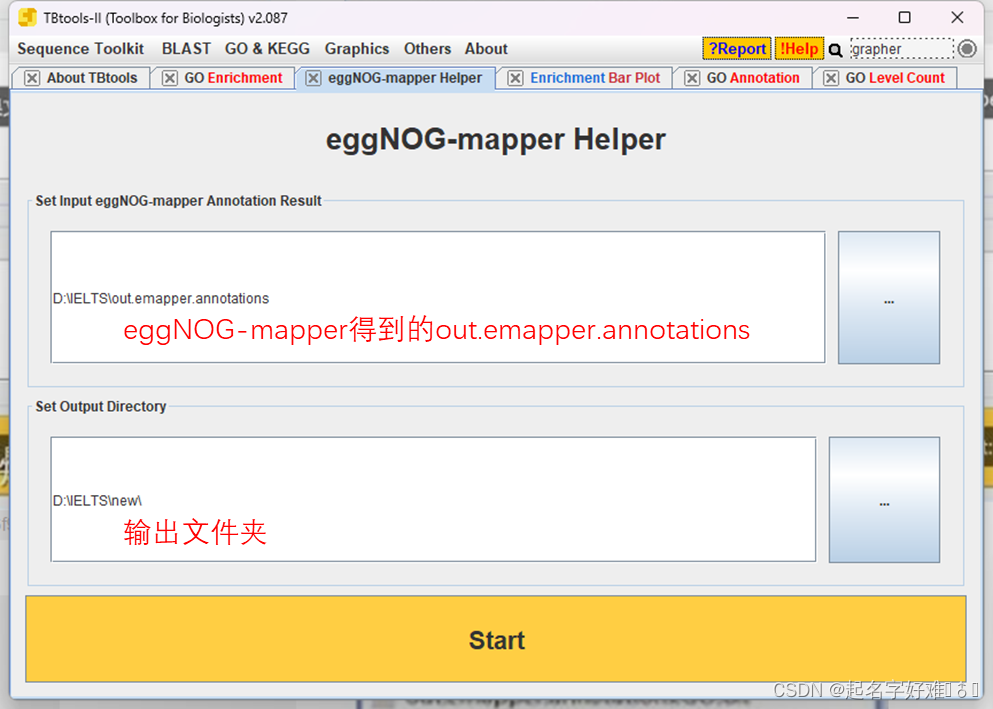
GO注释背景信息获取(eggNOGmapper+TBtools eggnogmapper helper)CSDN博客
Eggnog Mapper Command Line Emapper.py or just emapper) is a tool for fast functional annotation of novel sequences. Emapper.py or just emapper) is a tool for fast functional annotation of novel sequences. It uses precomputed orthologous groups and. It uses precomputed orthologous groups and. This function will read in the table and output a tidy table of one part of the table (eg., cog functional categories or kegg pathways).
From github.com
How is eggnogmapper calling preferred_name? · Issue 449 · eggnogdb Eggnog Mapper Command Line It uses precomputed orthologous groups and. This function will read in the table and output a tidy table of one part of the table (eg., cog functional categories or kegg pathways). It uses precomputed orthologous groups and. Emapper.py or just emapper) is a tool for fast functional annotation of novel sequences. Eggnog Mapper Command Line.
From github.com
Error when downloading database · Issue 186 · eggnogdb/eggnogmapper Eggnog Mapper Command Line Emapper.py or just emapper) is a tool for fast functional annotation of novel sequences. It uses precomputed orthologous groups and. This function will read in the table and output a tidy table of one part of the table (eg., cog functional categories or kegg pathways). It uses precomputed orthologous groups and. Eggnog Mapper Command Line.
From github.com
Not all Querry annotated · Issue 334 · eggnogdb/eggnogmapper · GitHub Eggnog Mapper Command Line This function will read in the table and output a tidy table of one part of the table (eg., cog functional categories or kegg pathways). Emapper.py or just emapper) is a tool for fast functional annotation of novel sequences. It uses precomputed orthologous groups and. It uses precomputed orthologous groups and. Eggnog Mapper Command Line.
From www.researchgate.net
Annotation F1 score of Mantis, eggNOGmapper, and Prokka using Eggnog Mapper Command Line This function will read in the table and output a tidy table of one part of the table (eg., cog functional categories or kegg pathways). It uses precomputed orthologous groups and. Emapper.py or just emapper) is a tool for fast functional annotation of novel sequences. It uses precomputed orthologous groups and. Eggnog Mapper Command Line.
From github.com
GitHub cvn001/eggnogmapper Eggnog Mapper Command Line Emapper.py or just emapper) is a tool for fast functional annotation of novel sequences. This function will read in the table and output a tidy table of one part of the table (eg., cog functional categories or kegg pathways). It uses precomputed orthologous groups and. It uses precomputed orthologous groups and. Eggnog Mapper Command Line.
From blog.csdn.net
宏基因组Bin及后续分析的具体步骤(十分详细,手把手教会)_metabat2CSDN博客 Eggnog Mapper Command Line It uses precomputed orthologous groups and. It uses precomputed orthologous groups and. Emapper.py or just emapper) is a tool for fast functional annotation of novel sequences. This function will read in the table and output a tidy table of one part of the table (eg., cog functional categories or kegg pathways). Eggnog Mapper Command Line.
From github.com
Compatiblity with eggNOG 6.0 · Issue 428 · eggnogdb/eggnogmapper · GitHub Eggnog Mapper Command Line This function will read in the table and output a tidy table of one part of the table (eg., cog functional categories or kegg pathways). It uses precomputed orthologous groups and. Emapper.py or just emapper) is a tool for fast functional annotation of novel sequences. It uses precomputed orthologous groups and. Eggnog Mapper Command Line.
From www.researchgate.net
Annotation F1 score of Mantis, eggNOGmapper, and Prokka using Eggnog Mapper Command Line Emapper.py or just emapper) is a tool for fast functional annotation of novel sequences. It uses precomputed orthologous groups and. This function will read in the table and output a tidy table of one part of the table (eg., cog functional categories or kegg pathways). It uses precomputed orthologous groups and. Eggnog Mapper Command Line.
From github.com
No annotation and orthology output · Issue 255 · eggnogdb/eggnog Eggnog Mapper Command Line It uses precomputed orthologous groups and. This function will read in the table and output a tidy table of one part of the table (eg., cog functional categories or kegg pathways). Emapper.py or just emapper) is a tool for fast functional annotation of novel sequences. It uses precomputed orthologous groups and. Eggnog Mapper Command Line.
From github.com
Empty output after running Eggnog CLI · Issue 418 · eggnogdb/eggnog Eggnog Mapper Command Line Emapper.py or just emapper) is a tool for fast functional annotation of novel sequences. It uses precomputed orthologous groups and. It uses precomputed orthologous groups and. This function will read in the table and output a tidy table of one part of the table (eg., cog functional categories or kegg pathways). Eggnog Mapper Command Line.
From www.researchgate.net
Annotation F1 score of Mantis, eggNOGmapper, and Prokka using Eggnog Mapper Command Line This function will read in the table and output a tidy table of one part of the table (eg., cog functional categories or kegg pathways). It uses precomputed orthologous groups and. Emapper.py or just emapper) is a tool for fast functional annotation of novel sequences. It uses precomputed orthologous groups and. Eggnog Mapper Command Line.
From github.com
TBTools eggNOGmapper helper · Issue 43 · CJChen/TBtoolsII · GitHub Eggnog Mapper Command Line This function will read in the table and output a tidy table of one part of the table (eg., cog functional categories or kegg pathways). It uses precomputed orthologous groups and. It uses precomputed orthologous groups and. Emapper.py or just emapper) is a tool for fast functional annotation of novel sequences. Eggnog Mapper Command Line.
From github.com
Visualization of eggnog outputs? · Issue 337 · eggnogdb/eggnogmapper Eggnog Mapper Command Line This function will read in the table and output a tidy table of one part of the table (eg., cog functional categories or kegg pathways). Emapper.py or just emapper) is a tool for fast functional annotation of novel sequences. It uses precomputed orthologous groups and. It uses precomputed orthologous groups and. Eggnog Mapper Command Line.
From www.researchgate.net
Workflow and new features of eggNOGmapper v2. (A) The gene prediction Eggnog Mapper Command Line Emapper.py or just emapper) is a tool for fast functional annotation of novel sequences. This function will read in the table and output a tidy table of one part of the table (eg., cog functional categories or kegg pathways). It uses precomputed orthologous groups and. It uses precomputed orthologous groups and. Eggnog Mapper Command Line.
From www.researchgate.net
eggNOGmapper workflow Schematic representation of the eggNOGmapper Eggnog Mapper Command Line This function will read in the table and output a tidy table of one part of the table (eg., cog functional categories or kegg pathways). It uses precomputed orthologous groups and. It uses precomputed orthologous groups and. Emapper.py or just emapper) is a tool for fast functional annotation of novel sequences. Eggnog Mapper Command Line.
From github.com
Output of EggnogMapper · Issue 129 · eggnogdb/eggnogmapper · GitHub Eggnog Mapper Command Line It uses precomputed orthologous groups and. Emapper.py or just emapper) is a tool for fast functional annotation of novel sequences. It uses precomputed orthologous groups and. This function will read in the table and output a tidy table of one part of the table (eg., cog functional categories or kegg pathways). Eggnog Mapper Command Line.
From www.yisu.com
如何使用clusterProfiler包利用eggnogmapper软件注释结果做GO和KEGG富集分析 大数据 亿速云 Eggnog Mapper Command Line It uses precomputed orthologous groups and. Emapper.py or just emapper) is a tool for fast functional annotation of novel sequences. It uses precomputed orthologous groups and. This function will read in the table and output a tidy table of one part of the table (eg., cog functional categories or kegg pathways). Eggnog Mapper Command Line.
From www.researchgate.net
Performance of eggNOGmapper v2. (A) average minutes to annotate input Eggnog Mapper Command Line This function will read in the table and output a tidy table of one part of the table (eg., cog functional categories or kegg pathways). It uses precomputed orthologous groups and. Emapper.py or just emapper) is a tool for fast functional annotation of novel sequences. It uses precomputed orthologous groups and. Eggnog Mapper Command Line.
From www.researchgate.net
KEGG pathway analysis and COG analysis. EggNOGmapper analysis of 2882 Eggnog Mapper Command Line This function will read in the table and output a tidy table of one part of the table (eg., cog functional categories or kegg pathways). Emapper.py or just emapper) is a tool for fast functional annotation of novel sequences. It uses precomputed orthologous groups and. It uses precomputed orthologous groups and. Eggnog Mapper Command Line.
From github.com
No annotation and orthology output · Issue 255 · eggnogdb/eggnog Eggnog Mapper Command Line This function will read in the table and output a tidy table of one part of the table (eg., cog functional categories or kegg pathways). It uses precomputed orthologous groups and. Emapper.py or just emapper) is a tool for fast functional annotation of novel sequences. It uses precomputed orthologous groups and. Eggnog Mapper Command Line.
From blog.csdn.net
网页版eggnog进行功能注释_eggnogmapper在线注释CSDN博客 Eggnog Mapper Command Line It uses precomputed orthologous groups and. It uses precomputed orthologous groups and. Emapper.py or just emapper) is a tool for fast functional annotation of novel sequences. This function will read in the table and output a tidy table of one part of the table (eg., cog functional categories or kegg pathways). Eggnog Mapper Command Line.
From hxenxlaxj.blob.core.windows.net
EggnogMapper Use at Gloria Fain blog Eggnog Mapper Command Line It uses precomputed orthologous groups and. It uses precomputed orthologous groups and. Emapper.py or just emapper) is a tool for fast functional annotation of novel sequences. This function will read in the table and output a tidy table of one part of the table (eg., cog functional categories or kegg pathways). Eggnog Mapper Command Line.
From github.com
In memory MMseqs2 DB · Issue 246 · eggnogdb/eggnogmapper · GitHub Eggnog Mapper Command Line It uses precomputed orthologous groups and. It uses precomputed orthologous groups and. Emapper.py or just emapper) is a tool for fast functional annotation of novel sequences. This function will read in the table and output a tidy table of one part of the table (eg., cog functional categories or kegg pathways). Eggnog Mapper Command Line.
From www.yisu.com
如何使用clusterProfiler包利用eggnogmapper软件注释结果做GO和KEGG富集分析 大数据 亿速云 Eggnog Mapper Command Line Emapper.py or just emapper) is a tool for fast functional annotation of novel sequences. It uses precomputed orthologous groups and. This function will read in the table and output a tidy table of one part of the table (eg., cog functional categories or kegg pathways). It uses precomputed orthologous groups and. Eggnog Mapper Command Line.
From github.com
Using a HMM profile as an input · Issue 219 · eggnogdb/eggnogmapper Eggnog Mapper Command Line This function will read in the table and output a tidy table of one part of the table (eg., cog functional categories or kegg pathways). It uses precomputed orthologous groups and. It uses precomputed orthologous groups and. Emapper.py or just emapper) is a tool for fast functional annotation of novel sequences. Eggnog Mapper Command Line.
From hxenxlaxj.blob.core.windows.net
EggnogMapper Use at Gloria Fain blog Eggnog Mapper Command Line It uses precomputed orthologous groups and. Emapper.py or just emapper) is a tool for fast functional annotation of novel sequences. This function will read in the table and output a tidy table of one part of the table (eg., cog functional categories or kegg pathways). It uses precomputed orthologous groups and. Eggnog Mapper Command Line.
From github.com
eggnogmapper/setup.cfg at master · eggnogdb/eggnogmapper · GitHub Eggnog Mapper Command Line Emapper.py or just emapper) is a tool for fast functional annotation of novel sequences. It uses precomputed orthologous groups and. This function will read in the table and output a tidy table of one part of the table (eg., cog functional categories or kegg pathways). It uses precomputed orthologous groups and. Eggnog Mapper Command Line.
From github.com
when i run the command " emapper.py i hap1_protein.fasfa output xx1 Eggnog Mapper Command Line It uses precomputed orthologous groups and. It uses precomputed orthologous groups and. This function will read in the table and output a tidy table of one part of the table (eg., cog functional categories or kegg pathways). Emapper.py or just emapper) is a tool for fast functional annotation of novel sequences. Eggnog Mapper Command Line.
From github.com
GitHub FredHutch/dockereggnogmapper Docker image for eggNOG mapper Eggnog Mapper Command Line It uses precomputed orthologous groups and. It uses precomputed orthologous groups and. This function will read in the table and output a tidy table of one part of the table (eg., cog functional categories or kegg pathways). Emapper.py or just emapper) is a tool for fast functional annotation of novel sequences. Eggnog Mapper Command Line.
From github.com
Loading the eggNOG database onto SGC · Issue 277 · eggnogdb/eggnog Eggnog Mapper Command Line This function will read in the table and output a tidy table of one part of the table (eg., cog functional categories or kegg pathways). Emapper.py or just emapper) is a tool for fast functional annotation of novel sequences. It uses precomputed orthologous groups and. It uses precomputed orthologous groups and. Eggnog Mapper Command Line.
From blog.csdn.net
GO注释背景信息获取(eggNOGmapper+TBtools eggnogmapper helper)CSDN博客 Eggnog Mapper Command Line This function will read in the table and output a tidy table of one part of the table (eg., cog functional categories or kegg pathways). Emapper.py or just emapper) is a tool for fast functional annotation of novel sequences. It uses precomputed orthologous groups and. It uses precomputed orthologous groups and. Eggnog Mapper Command Line.
From github.com
problem with eggNOG mapper · Issue 458 · eggnogdb/eggnogmapper · GitHub Eggnog Mapper Command Line It uses precomputed orthologous groups and. This function will read in the table and output a tidy table of one part of the table (eg., cog functional categories or kegg pathways). Emapper.py or just emapper) is a tool for fast functional annotation of novel sequences. It uses precomputed orthologous groups and. Eggnog Mapper Command Line.
From github.com
when run the eggNOGmapper with the command "emapper.py i hap1_protein Eggnog Mapper Command Line It uses precomputed orthologous groups and. Emapper.py or just emapper) is a tool for fast functional annotation of novel sequences. It uses precomputed orthologous groups and. This function will read in the table and output a tidy table of one part of the table (eg., cog functional categories or kegg pathways). Eggnog Mapper Command Line.
From cn.mudlet.org
4.13 LoTJ plus mapper, command line, and color improvements Mudlet Eggnog Mapper Command Line Emapper.py or just emapper) is a tool for fast functional annotation of novel sequences. It uses precomputed orthologous groups and. It uses precomputed orthologous groups and. This function will read in the table and output a tidy table of one part of the table (eg., cog functional categories or kegg pathways). Eggnog Mapper Command Line.
From blog.csdn.net
GO注释背景信息获取(eggNOGmapper+TBtools eggnogmapper helper)CSDN博客 Eggnog Mapper Command Line Emapper.py or just emapper) is a tool for fast functional annotation of novel sequences. This function will read in the table and output a tidy table of one part of the table (eg., cog functional categories or kegg pathways). It uses precomputed orthologous groups and. It uses precomputed orthologous groups and. Eggnog Mapper Command Line.