Adapter Ligation Rna Seq . Findadapt is a software tool that can automatically identify adapter patterns in small rna sequencing data without relying on prior information. An ngs experiment requires the. It can handle diverse read structures. First, we demonstrate how to optimize the three variables (dmso, peg and adapter. Small rna with 5' recessed ends are poor substrates for enzymatic adapter ligation, but this 5' adapter ligation problem can go undetected if the library. Next generation sequencing (ngs) is the most common technique for studying small rna expression. The ligation of dna oligonucleotide sequencing adapters to unknown rna allows the rna to be sequenced via its cdna after reverse.
from www.labome.com
Findadapt is a software tool that can automatically identify adapter patterns in small rna sequencing data without relying on prior information. Small rna with 5' recessed ends are poor substrates for enzymatic adapter ligation, but this 5' adapter ligation problem can go undetected if the library. The ligation of dna oligonucleotide sequencing adapters to unknown rna allows the rna to be sequenced via its cdna after reverse. It can handle diverse read structures. An ngs experiment requires the. Next generation sequencing (ngs) is the most common technique for studying small rna expression. First, we demonstrate how to optimize the three variables (dmso, peg and adapter.
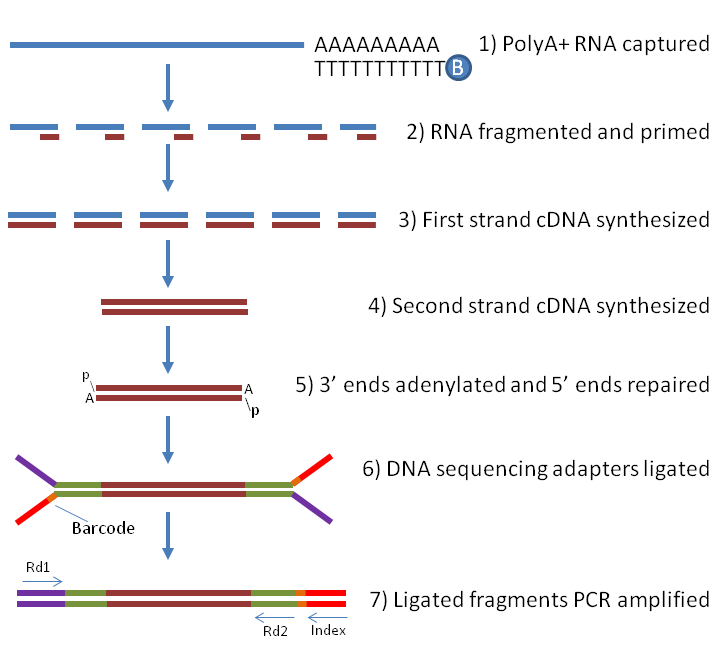
RNAseq Using Next Generation Sequencing
Adapter Ligation Rna Seq Next generation sequencing (ngs) is the most common technique for studying small rna expression. It can handle diverse read structures. Next generation sequencing (ngs) is the most common technique for studying small rna expression. Findadapt is a software tool that can automatically identify adapter patterns in small rna sequencing data without relying on prior information. The ligation of dna oligonucleotide sequencing adapters to unknown rna allows the rna to be sequenced via its cdna after reverse. Small rna with 5' recessed ends are poor substrates for enzymatic adapter ligation, but this 5' adapter ligation problem can go undetected if the library. First, we demonstrate how to optimize the three variables (dmso, peg and adapter. An ngs experiment requires the.
From www.lubio.ch
What are NGS adapters? Different structures and features Adapter Ligation Rna Seq First, we demonstrate how to optimize the three variables (dmso, peg and adapter. It can handle diverse read structures. The ligation of dna oligonucleotide sequencing adapters to unknown rna allows the rna to be sequenced via its cdna after reverse. Findadapt is a software tool that can automatically identify adapter patterns in small rna sequencing data without relying on prior. Adapter Ligation Rna Seq.
From www.researchgate.net
Validation of key enzymatic steps. (A) Ligation of the 3' adapter in Adapter Ligation Rna Seq First, we demonstrate how to optimize the three variables (dmso, peg and adapter. Small rna with 5' recessed ends are poor substrates for enzymatic adapter ligation, but this 5' adapter ligation problem can go undetected if the library. Findadapt is a software tool that can automatically identify adapter patterns in small rna sequencing data without relying on prior information. Next. Adapter Ligation Rna Seq.
From www.researchgate.net
Synthetic RNA ligation to 3′ adapter is enhanced by using a pool of 3 Adapter Ligation Rna Seq The ligation of dna oligonucleotide sequencing adapters to unknown rna allows the rna to be sequenced via its cdna after reverse. Next generation sequencing (ngs) is the most common technique for studying small rna expression. Small rna with 5' recessed ends are poor substrates for enzymatic adapter ligation, but this 5' adapter ligation problem can go undetected if the library.. Adapter Ligation Rna Seq.
From www.researchgate.net
Key differences among different commercially available library Adapter Ligation Rna Seq First, we demonstrate how to optimize the three variables (dmso, peg and adapter. Small rna with 5' recessed ends are poor substrates for enzymatic adapter ligation, but this 5' adapter ligation problem can go undetected if the library. It can handle diverse read structures. Findadapt is a software tool that can automatically identify adapter patterns in small rna sequencing data. Adapter Ligation Rna Seq.
From www.youtube.com
What is RNA seq? Part 2 Y Adapter Ligation Bridge PCR Adapter Ligation Rna Seq The ligation of dna oligonucleotide sequencing adapters to unknown rna allows the rna to be sequenced via its cdna after reverse. Findadapt is a software tool that can automatically identify adapter patterns in small rna sequencing data without relying on prior information. Next generation sequencing (ngs) is the most common technique for studying small rna expression. First, we demonstrate how. Adapter Ligation Rna Seq.
From www.sigmaaldrich.cn
Small RNA and miRNA (microRNA) Sequencing Adapter Ligation Rna Seq The ligation of dna oligonucleotide sequencing adapters to unknown rna allows the rna to be sequenced via its cdna after reverse. An ngs experiment requires the. Findadapt is a software tool that can automatically identify adapter patterns in small rna sequencing data without relying on prior information. First, we demonstrate how to optimize the three variables (dmso, peg and adapter.. Adapter Ligation Rna Seq.
From www.researchgate.net
Adaptor ligation is followed by sizeselection and PCR. Download Adapter Ligation Rna Seq An ngs experiment requires the. It can handle diverse read structures. First, we demonstrate how to optimize the three variables (dmso, peg and adapter. Findadapt is a software tool that can automatically identify adapter patterns in small rna sequencing data without relying on prior information. The ligation of dna oligonucleotide sequencing adapters to unknown rna allows the rna to be. Adapter Ligation Rna Seq.
From www.rna-seqblog.com
SmallRNA Sequencing of Single Cells RNASeq Blog Adapter Ligation Rna Seq Next generation sequencing (ngs) is the most common technique for studying small rna expression. First, we demonstrate how to optimize the three variables (dmso, peg and adapter. Findadapt is a software tool that can automatically identify adapter patterns in small rna sequencing data without relying on prior information. Small rna with 5' recessed ends are poor substrates for enzymatic adapter. Adapter Ligation Rna Seq.
From www.researchgate.net
Adapter ligation to 5 9 ends of RNA templates with different Adapter Ligation Rna Seq It can handle diverse read structures. Findadapt is a software tool that can automatically identify adapter patterns in small rna sequencing data without relying on prior information. Next generation sequencing (ngs) is the most common technique for studying small rna expression. An ngs experiment requires the. Small rna with 5' recessed ends are poor substrates for enzymatic adapter ligation, but. Adapter Ligation Rna Seq.
From www.activemotif.com
YourSeq (Full Transcript & 3’DGE) RNA Sequencing Services Adapter Ligation Rna Seq Findadapt is a software tool that can automatically identify adapter patterns in small rna sequencing data without relying on prior information. The ligation of dna oligonucleotide sequencing adapters to unknown rna allows the rna to be sequenced via its cdna after reverse. Small rna with 5' recessed ends are poor substrates for enzymatic adapter ligation, but this 5' adapter ligation. Adapter Ligation Rna Seq.
From www.frontiersin.org
Frontiers A HighThroughput Method for Illumina RNASeq Library Adapter Ligation Rna Seq Small rna with 5' recessed ends are poor substrates for enzymatic adapter ligation, but this 5' adapter ligation problem can go undetected if the library. Findadapt is a software tool that can automatically identify adapter patterns in small rna sequencing data without relying on prior information. The ligation of dna oligonucleotide sequencing adapters to unknown rna allows the rna to. Adapter Ligation Rna Seq.
From www.labome.com
RNAseq Using Next Generation Sequencing Adapter Ligation Rna Seq Small rna with 5' recessed ends are poor substrates for enzymatic adapter ligation, but this 5' adapter ligation problem can go undetected if the library. Next generation sequencing (ngs) is the most common technique for studying small rna expression. It can handle diverse read structures. First, we demonstrate how to optimize the three variables (dmso, peg and adapter. Findadapt is. Adapter Ligation Rna Seq.
From www.bioz.com
Adapter Ligated Rna Thermo Fisher Bioz Adapter Ligation Rna Seq Next generation sequencing (ngs) is the most common technique for studying small rna expression. Findadapt is a software tool that can automatically identify adapter patterns in small rna sequencing data without relying on prior information. Small rna with 5' recessed ends are poor substrates for enzymatic adapter ligation, but this 5' adapter ligation problem can go undetected if the library.. Adapter Ligation Rna Seq.
From tucf-genomics.tufts.edu
TUFTS TUCF Genomics Adapter Ligation Rna Seq The ligation of dna oligonucleotide sequencing adapters to unknown rna allows the rna to be sequenced via its cdna after reverse. It can handle diverse read structures. First, we demonstrate how to optimize the three variables (dmso, peg and adapter. Small rna with 5' recessed ends are poor substrates for enzymatic adapter ligation, but this 5' adapter ligation problem can. Adapter Ligation Rna Seq.
From www.researchgate.net
Original twoadaptor ligation protocol for small RNAseq analysis Adapter Ligation Rna Seq Next generation sequencing (ngs) is the most common technique for studying small rna expression. An ngs experiment requires the. Small rna with 5' recessed ends are poor substrates for enzymatic adapter ligation, but this 5' adapter ligation problem can go undetected if the library. The ligation of dna oligonucleotide sequencing adapters to unknown rna allows the rna to be sequenced. Adapter Ligation Rna Seq.
From www.researchgate.net
Improved twoadaptor ligationbased methods for small RNAseq analysis Adapter Ligation Rna Seq Small rna with 5' recessed ends are poor substrates for enzymatic adapter ligation, but this 5' adapter ligation problem can go undetected if the library. An ngs experiment requires the. Next generation sequencing (ngs) is the most common technique for studying small rna expression. Findadapt is a software tool that can automatically identify adapter patterns in small rna sequencing data. Adapter Ligation Rna Seq.
From www.researchgate.net
Ligation schemes for sequencing library preparation. a Twoadapter Adapter Ligation Rna Seq It can handle diverse read structures. Next generation sequencing (ngs) is the most common technique for studying small rna expression. An ngs experiment requires the. First, we demonstrate how to optimize the three variables (dmso, peg and adapter. Findadapt is a software tool that can automatically identify adapter patterns in small rna sequencing data without relying on prior information. The. Adapter Ligation Rna Seq.
From www.researchgate.net
Original twoadaptor ligation protocol for small RNAseq analysis Adapter Ligation Rna Seq Findadapt is a software tool that can automatically identify adapter patterns in small rna sequencing data without relying on prior information. It can handle diverse read structures. First, we demonstrate how to optimize the three variables (dmso, peg and adapter. The ligation of dna oligonucleotide sequencing adapters to unknown rna allows the rna to be sequenced via its cdna after. Adapter Ligation Rna Seq.
From www.researchgate.net
Methods for strandspecific RNASeq. (a) Ligation of the 3 0 Adapter Ligation Rna Seq Small rna with 5' recessed ends are poor substrates for enzymatic adapter ligation, but this 5' adapter ligation problem can go undetected if the library. An ngs experiment requires the. Next generation sequencing (ngs) is the most common technique for studying small rna expression. The ligation of dna oligonucleotide sequencing adapters to unknown rna allows the rna to be sequenced. Adapter Ligation Rna Seq.
From www.rna-seqblog.com
Accurate adapter information is crucial for reproducibility and Adapter Ligation Rna Seq Small rna with 5' recessed ends are poor substrates for enzymatic adapter ligation, but this 5' adapter ligation problem can go undetected if the library. It can handle diverse read structures. An ngs experiment requires the. First, we demonstrate how to optimize the three variables (dmso, peg and adapter. The ligation of dna oligonucleotide sequencing adapters to unknown rna allows. Adapter Ligation Rna Seq.
From stagepages.idtdna.com
xGen RNA Library Prep Kit IDT Adapter Ligation Rna Seq First, we demonstrate how to optimize the three variables (dmso, peg and adapter. Findadapt is a software tool that can automatically identify adapter patterns in small rna sequencing data without relying on prior information. It can handle diverse read structures. Small rna with 5' recessed ends are poor substrates for enzymatic adapter ligation, but this 5' adapter ligation problem can. Adapter Ligation Rna Seq.
From genome.cshlp.org
Strandspecific deep sequencing of the transcriptome Adapter Ligation Rna Seq Small rna with 5' recessed ends are poor substrates for enzymatic adapter ligation, but this 5' adapter ligation problem can go undetected if the library. Findadapt is a software tool that can automatically identify adapter patterns in small rna sequencing data without relying on prior information. An ngs experiment requires the. It can handle diverse read structures. Next generation sequencing. Adapter Ligation Rna Seq.
From www.researchgate.net
Library preparation (A) Sequence of adapters (single index) used in our Adapter Ligation Rna Seq First, we demonstrate how to optimize the three variables (dmso, peg and adapter. The ligation of dna oligonucleotide sequencing adapters to unknown rna allows the rna to be sequenced via its cdna after reverse. Findadapt is a software tool that can automatically identify adapter patterns in small rna sequencing data without relying on prior information. Next generation sequencing (ngs) is. Adapter Ligation Rna Seq.
From www.researchgate.net
Targeted RNASeq by target capture. (a) The CaptureSeq method is Adapter Ligation Rna Seq The ligation of dna oligonucleotide sequencing adapters to unknown rna allows the rna to be sequenced via its cdna after reverse. Next generation sequencing (ngs) is the most common technique for studying small rna expression. First, we demonstrate how to optimize the three variables (dmso, peg and adapter. Small rna with 5' recessed ends are poor substrates for enzymatic adapter. Adapter Ligation Rna Seq.
From www.researchgate.net
Schematic of RNAseq library generation protocols. RNA fragments are Adapter Ligation Rna Seq Small rna with 5' recessed ends are poor substrates for enzymatic adapter ligation, but this 5' adapter ligation problem can go undetected if the library. First, we demonstrate how to optimize the three variables (dmso, peg and adapter. Findadapt is a software tool that can automatically identify adapter patterns in small rna sequencing data without relying on prior information. The. Adapter Ligation Rna Seq.
From www.researchgate.net
Method overview. Step 1 Ligation. RNA, shown in blue, is ligated to a Adapter Ligation Rna Seq It can handle diverse read structures. First, we demonstrate how to optimize the three variables (dmso, peg and adapter. Next generation sequencing (ngs) is the most common technique for studying small rna expression. Findadapt is a software tool that can automatically identify adapter patterns in small rna sequencing data without relying on prior information. The ligation of dna oligonucleotide sequencing. Adapter Ligation Rna Seq.
From zymoresearch.eu
What is RNA Seq and Why Use RNA Sequencing? Zymo Research International Adapter Ligation Rna Seq Next generation sequencing (ngs) is the most common technique for studying small rna expression. It can handle diverse read structures. The ligation of dna oligonucleotide sequencing adapters to unknown rna allows the rna to be sequenced via its cdna after reverse. An ngs experiment requires the. Small rna with 5' recessed ends are poor substrates for enzymatic adapter ligation, but. Adapter Ligation Rna Seq.
From www.researchgate.net
DNA hairpin adapter ligation. (A) DNA hairpin adapter for LOTTEseq Adapter Ligation Rna Seq Small rna with 5' recessed ends are poor substrates for enzymatic adapter ligation, but this 5' adapter ligation problem can go undetected if the library. First, we demonstrate how to optimize the three variables (dmso, peg and adapter. It can handle diverse read structures. Next generation sequencing (ngs) is the most common technique for studying small rna expression. The ligation. Adapter Ligation Rna Seq.
From www.researchgate.net
Data production workflow for RNASeq. RNASeq requires building Adapter Ligation Rna Seq Findadapt is a software tool that can automatically identify adapter patterns in small rna sequencing data without relying on prior information. Small rna with 5' recessed ends are poor substrates for enzymatic adapter ligation, but this 5' adapter ligation problem can go undetected if the library. First, we demonstrate how to optimize the three variables (dmso, peg and adapter. An. Adapter Ligation Rna Seq.
From www.bioz.com
Adapter Ligated Rna Thermo Fisher Bioz Adapter Ligation Rna Seq Small rna with 5' recessed ends are poor substrates for enzymatic adapter ligation, but this 5' adapter ligation problem can go undetected if the library. It can handle diverse read structures. Next generation sequencing (ngs) is the most common technique for studying small rna expression. Findadapt is a software tool that can automatically identify adapter patterns in small rna sequencing. Adapter Ligation Rna Seq.
From www.researchgate.net
The 5' sequencing adapter ligation problem with hairpin RNA. A Adapter Ligation Rna Seq Small rna with 5' recessed ends are poor substrates for enzymatic adapter ligation, but this 5' adapter ligation problem can go undetected if the library. Next generation sequencing (ngs) is the most common technique for studying small rna expression. Findadapt is a software tool that can automatically identify adapter patterns in small rna sequencing data without relying on prior information.. Adapter Ligation Rna Seq.
From www.researchgate.net
DNA hairpin adapter ligation. (A) DNA hairpin adapter for LOTTEseq Adapter Ligation Rna Seq It can handle diverse read structures. First, we demonstrate how to optimize the three variables (dmso, peg and adapter. Small rna with 5' recessed ends are poor substrates for enzymatic adapter ligation, but this 5' adapter ligation problem can go undetected if the library. The ligation of dna oligonucleotide sequencing adapters to unknown rna allows the rna to be sequenced. Adapter Ligation Rna Seq.
From genxpro.net
TrueQuant SmallRNA Seq Kit Ultra Low Input Single Tube Adapter Ligation Rna Seq The ligation of dna oligonucleotide sequencing adapters to unknown rna allows the rna to be sequenced via its cdna after reverse. It can handle diverse read structures. First, we demonstrate how to optimize the three variables (dmso, peg and adapter. Findadapt is a software tool that can automatically identify adapter patterns in small rna sequencing data without relying on prior. Adapter Ligation Rna Seq.
From www.researchgate.net
Ion Total RNASeq approach. The protocol for RNA library preparation Adapter Ligation Rna Seq It can handle diverse read structures. Next generation sequencing (ngs) is the most common technique for studying small rna expression. An ngs experiment requires the. The ligation of dna oligonucleotide sequencing adapters to unknown rna allows the rna to be sequenced via its cdna after reverse. Findadapt is a software tool that can automatically identify adapter patterns in small rna. Adapter Ligation Rna Seq.
From www.researchgate.net
Original twoadaptor ligation protocol for small RNAseq analysis Adapter Ligation Rna Seq An ngs experiment requires the. First, we demonstrate how to optimize the three variables (dmso, peg and adapter. The ligation of dna oligonucleotide sequencing adapters to unknown rna allows the rna to be sequenced via its cdna after reverse. Findadapt is a software tool that can automatically identify adapter patterns in small rna sequencing data without relying on prior information.. Adapter Ligation Rna Seq.