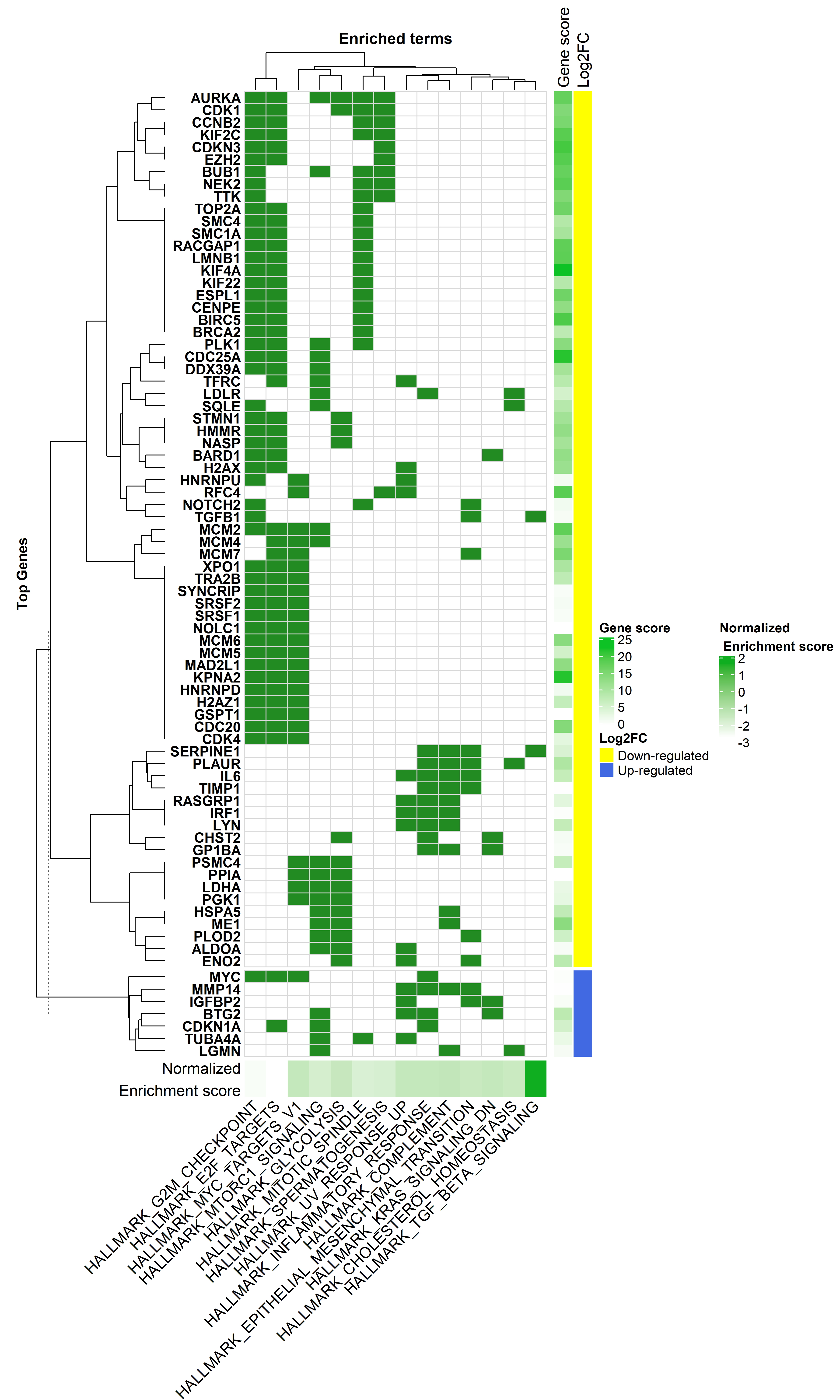
Gene Set Enrichment Analysis David . gene set enrichment analysis. david bioinformatics provides a comprehensive set of tools to understand the biological meaning behind large lists of. A common approach to functional genomics data is gene enrichment analysis based on the. learn how to use david tools to analyze gene lists and get functional annotation, enrichment, clustering, and classification. — david is a bioinformatics resource system for functional annotation and enrichment analysis of gene lists. — david is a popular bioinformatics resource system including a web server and web service for functional. Learn about its history, features, api,. Find out the features, options, and. david is a database and tool for extracting biological meaning from large gene/protein lists.
from www.biostars.org
Find out the features, options, and. david is a database and tool for extracting biological meaning from large gene/protein lists. — david is a popular bioinformatics resource system including a web server and web service for functional. gene set enrichment analysis. david bioinformatics provides a comprehensive set of tools to understand the biological meaning behind large lists of. Learn about its history, features, api,. A common approach to functional genomics data is gene enrichment analysis based on the. — david is a bioinformatics resource system for functional annotation and enrichment analysis of gene lists. learn how to use david tools to analyze gene lists and get functional annotation, enrichment, clustering, and classification.
Gene Set Enrichment Analysis
Gene Set Enrichment Analysis David — david is a popular bioinformatics resource system including a web server and web service for functional. — david is a bioinformatics resource system for functional annotation and enrichment analysis of gene lists. — david is a popular bioinformatics resource system including a web server and web service for functional. gene set enrichment analysis. Learn about its history, features, api,. learn how to use david tools to analyze gene lists and get functional annotation, enrichment, clustering, and classification. david is a database and tool for extracting biological meaning from large gene/protein lists. A common approach to functional genomics data is gene enrichment analysis based on the. david bioinformatics provides a comprehensive set of tools to understand the biological meaning behind large lists of. Find out the features, options, and.
From www.g2nb.org
Gene Set Enrichment Analysis g2nb Gene Set Enrichment Analysis David gene set enrichment analysis. david bioinformatics provides a comprehensive set of tools to understand the biological meaning behind large lists of. Learn about its history, features, api,. learn how to use david tools to analyze gene lists and get functional annotation, enrichment, clustering, and classification. — david is a popular bioinformatics resource system including a web. Gene Set Enrichment Analysis David.
From www.pnas.org
Gene set enrichment analysis A knowledgebased approach for Gene Set Enrichment Analysis David learn how to use david tools to analyze gene lists and get functional annotation, enrichment, clustering, and classification. david is a database and tool for extracting biological meaning from large gene/protein lists. gene set enrichment analysis. A common approach to functional genomics data is gene enrichment analysis based on the. Find out the features, options, and. . Gene Set Enrichment Analysis David.
From www.slideserve.com
PPT Gene Set Enrichment Analysis PowerPoint Presentation, free Gene Set Enrichment Analysis David david bioinformatics provides a comprehensive set of tools to understand the biological meaning behind large lists of. learn how to use david tools to analyze gene lists and get functional annotation, enrichment, clustering, and classification. Learn about its history, features, api,. — david is a popular bioinformatics resource system including a web server and web service for. Gene Set Enrichment Analysis David.
From bigomics.ch
How to Perform Gene Set Enrichment Analysis Gene Set Enrichment Analysis David learn how to use david tools to analyze gene lists and get functional annotation, enrichment, clustering, and classification. david bioinformatics provides a comprehensive set of tools to understand the biological meaning behind large lists of. Learn about its history, features, api,. — david is a popular bioinformatics resource system including a web server and web service for. Gene Set Enrichment Analysis David.
From www.researchgate.net
The gene set enrichment analysis (GSEA) and consensus clustering of Gene Set Enrichment Analysis David — david is a popular bioinformatics resource system including a web server and web service for functional. — david is a bioinformatics resource system for functional annotation and enrichment analysis of gene lists. gene set enrichment analysis. Learn about its history, features, api,. david is a database and tool for extracting biological meaning from large gene/protein. Gene Set Enrichment Analysis David.
From www.researchgate.net
Gene Set Enrichment Analysis (GSEA) for Gene Ontology (GO) and Kyoto Gene Set Enrichment Analysis David gene set enrichment analysis. Find out the features, options, and. Learn about its history, features, api,. — david is a popular bioinformatics resource system including a web server and web service for functional. A common approach to functional genomics data is gene enrichment analysis based on the. — david is a bioinformatics resource system for functional annotation. Gene Set Enrichment Analysis David.
From www.researchgate.net
Gene set enrichment analysis identifies E2F and H3K27me3 signature Gene Set Enrichment Analysis David learn how to use david tools to analyze gene lists and get functional annotation, enrichment, clustering, and classification. A common approach to functional genomics data is gene enrichment analysis based on the. Find out the features, options, and. Learn about its history, features, api,. — david is a bioinformatics resource system for functional annotation and enrichment analysis of. Gene Set Enrichment Analysis David.
From www.researchgate.net
DAVID GO functional category enrichment analysis. Gene ontology (GO Gene Set Enrichment Analysis David A common approach to functional genomics data is gene enrichment analysis based on the. david is a database and tool for extracting biological meaning from large gene/protein lists. Learn about its history, features, api,. david bioinformatics provides a comprehensive set of tools to understand the biological meaning behind large lists of. gene set enrichment analysis. learn. Gene Set Enrichment Analysis David.
From www.biostars.org
Clustering of DAVID gene enrichment results from gene expression studies Gene Set Enrichment Analysis David — david is a bioinformatics resource system for functional annotation and enrichment analysis of gene lists. david bioinformatics provides a comprehensive set of tools to understand the biological meaning behind large lists of. A common approach to functional genomics data is gene enrichment analysis based on the. learn how to use david tools to analyze gene lists. Gene Set Enrichment Analysis David.
From www.researchgate.net
DAVID enrichment results of differentially expressed genes. (A Gene Set Enrichment Analysis David — david is a popular bioinformatics resource system including a web server and web service for functional. gene set enrichment analysis. david bioinformatics provides a comprehensive set of tools to understand the biological meaning behind large lists of. Learn about its history, features, api,. david is a database and tool for extracting biological meaning from large. Gene Set Enrichment Analysis David.
From www.researchgate.net
Gene set enrichment analysis of GRAMD1a and GRAMD2a indicated distinct Gene Set Enrichment Analysis David david is a database and tool for extracting biological meaning from large gene/protein lists. Learn about its history, features, api,. A common approach to functional genomics data is gene enrichment analysis based on the. david bioinformatics provides a comprehensive set of tools to understand the biological meaning behind large lists of. — david is a popular bioinformatics. Gene Set Enrichment Analysis David.
From www.researchgate.net
Gene Ontology Enrichment Analysis performed using David Bioinformatics Gene Set Enrichment Analysis David Learn about its history, features, api,. david is a database and tool for extracting biological meaning from large gene/protein lists. david bioinformatics provides a comprehensive set of tools to understand the biological meaning behind large lists of. — david is a popular bioinformatics resource system including a web server and web service for functional. learn how. Gene Set Enrichment Analysis David.
From www.researchgate.net
Gene ontology enrichment analyses by DAVID using the 41 identified Gene Set Enrichment Analysis David gene set enrichment analysis. Learn about its history, features, api,. david is a database and tool for extracting biological meaning from large gene/protein lists. learn how to use david tools to analyze gene lists and get functional annotation, enrichment, clustering, and classification. — david is a bioinformatics resource system for functional annotation and enrichment analysis of. Gene Set Enrichment Analysis David.
From www.researchgate.net
Gene Ontology (GO) enrichment analysis for lncRNAs by DAVID Gene Set Enrichment Analysis David Learn about its history, features, api,. — david is a popular bioinformatics resource system including a web server and web service for functional. learn how to use david tools to analyze gene lists and get functional annotation, enrichment, clustering, and classification. david is a database and tool for extracting biological meaning from large gene/protein lists. david. Gene Set Enrichment Analysis David.
From www.researchgate.net
Gene set enrichment analysis (GSEA) and gene set variation Gene Set Enrichment Analysis David Learn about its history, features, api,. — david is a popular bioinformatics resource system including a web server and web service for functional. A common approach to functional genomics data is gene enrichment analysis based on the. — david is a bioinformatics resource system for functional annotation and enrichment analysis of gene lists. Find out the features, options,. Gene Set Enrichment Analysis David.
From www.biostars.org
Gene Set Enrichment Analysis Gene Set Enrichment Analysis David Find out the features, options, and. learn how to use david tools to analyze gene lists and get functional annotation, enrichment, clustering, and classification. A common approach to functional genomics data is gene enrichment analysis based on the. — david is a popular bioinformatics resource system including a web server and web service for functional. david bioinformatics. Gene Set Enrichment Analysis David.
From www.slideserve.com
PPT Gene Set Analysis using R and Bioconductor PowerPoint Gene Set Enrichment Analysis David A common approach to functional genomics data is gene enrichment analysis based on the. david is a database and tool for extracting biological meaning from large gene/protein lists. Learn about its history, features, api,. Find out the features, options, and. — david is a popular bioinformatics resource system including a web server and web service for functional. . Gene Set Enrichment Analysis David.
From www.researchgate.net
Gene sets enrichment analysis and proteinprotein interaction analysis Gene Set Enrichment Analysis David — david is a bioinformatics resource system for functional annotation and enrichment analysis of gene lists. gene set enrichment analysis. david is a database and tool for extracting biological meaning from large gene/protein lists. Find out the features, options, and. — david is a popular bioinformatics resource system including a web server and web service for. Gene Set Enrichment Analysis David.
From www.researchgate.net
(A) Gene set enrichment analysis (GSEA) of the most upregulated and Gene Set Enrichment Analysis David Learn about its history, features, api,. david is a database and tool for extracting biological meaning from large gene/protein lists. gene set enrichment analysis. Find out the features, options, and. learn how to use david tools to analyze gene lists and get functional annotation, enrichment, clustering, and classification. — david is a popular bioinformatics resource system. Gene Set Enrichment Analysis David.
From crazyhottommy.blogspot.com
Diving into and Genomics Gene Set Enrichment Analysis (GSEA Gene Set Enrichment Analysis David learn how to use david tools to analyze gene lists and get functional annotation, enrichment, clustering, and classification. A common approach to functional genomics data is gene enrichment analysis based on the. — david is a popular bioinformatics resource system including a web server and web service for functional. Find out the features, options, and. — david. Gene Set Enrichment Analysis David.
From www.researchgate.net
Gene set enrichment analysis (GSEA) of genes highly expressed in Gene Set Enrichment Analysis David david bioinformatics provides a comprehensive set of tools to understand the biological meaning behind large lists of. A common approach to functional genomics data is gene enrichment analysis based on the. — david is a popular bioinformatics resource system including a web server and web service for functional. Find out the features, options, and. Learn about its history,. Gene Set Enrichment Analysis David.
From www.researchgate.net
Comparison between DAVID Gene Set Enrichment Analysis and Pathway Gene Set Enrichment Analysis David — david is a bioinformatics resource system for functional annotation and enrichment analysis of gene lists. Find out the features, options, and. — david is a popular bioinformatics resource system including a web server and web service for functional. A common approach to functional genomics data is gene enrichment analysis based on the. gene set enrichment analysis.. Gene Set Enrichment Analysis David.
From www.omicsonline.org
Gene set enrichment analysis Gene Set Enrichment Analysis David gene set enrichment analysis. learn how to use david tools to analyze gene lists and get functional annotation, enrichment, clustering, and classification. david is a database and tool for extracting biological meaning from large gene/protein lists. — david is a popular bioinformatics resource system including a web server and web service for functional. Find out the. Gene Set Enrichment Analysis David.
From www.researchgate.net
DAVID functional enrichment analysis, functional Gene Ontology analysis Gene Set Enrichment Analysis David david is a database and tool for extracting biological meaning from large gene/protein lists. david bioinformatics provides a comprehensive set of tools to understand the biological meaning behind large lists of. Find out the features, options, and. gene set enrichment analysis. — david is a popular bioinformatics resource system including a web server and web service. Gene Set Enrichment Analysis David.
From www.researchgate.net
The results of gene set enrichment analysis (GSEA). (A) The top three Gene Set Enrichment Analysis David david is a database and tool for extracting biological meaning from large gene/protein lists. Find out the features, options, and. Learn about its history, features, api,. david bioinformatics provides a comprehensive set of tools to understand the biological meaning behind large lists of. A common approach to functional genomics data is gene enrichment analysis based on the. . Gene Set Enrichment Analysis David.
From www.researchgate.net
Gene set enrichment analysis (GSEA) showing an immune system remolding Gene Set Enrichment Analysis David Learn about its history, features, api,. — david is a bioinformatics resource system for functional annotation and enrichment analysis of gene lists. — david is a popular bioinformatics resource system including a web server and web service for functional. david bioinformatics provides a comprehensive set of tools to understand the biological meaning behind large lists of. . Gene Set Enrichment Analysis David.
From www.researchgate.net
Gene set enrichment analysis and gene coexpression networks identify Gene Set Enrichment Analysis David gene set enrichment analysis. — david is a popular bioinformatics resource system including a web server and web service for functional. learn how to use david tools to analyze gene lists and get functional annotation, enrichment, clustering, and classification. — david is a bioinformatics resource system for functional annotation and enrichment analysis of gene lists. Learn. Gene Set Enrichment Analysis David.
From www.researchgate.net
Gene set enrichment analysis for KEGG pathways mapping showing Gene Set Enrichment Analysis David david is a database and tool for extracting biological meaning from large gene/protein lists. A common approach to functional genomics data is gene enrichment analysis based on the. Learn about its history, features, api,. learn how to use david tools to analyze gene lists and get functional annotation, enrichment, clustering, and classification. gene set enrichment analysis. . Gene Set Enrichment Analysis David.
From www.researchgate.net
Gene set enrichment analysis. Gene set enrichment analysis of models Gene Set Enrichment Analysis David — david is a bioinformatics resource system for functional annotation and enrichment analysis of gene lists. gene set enrichment analysis. david bioinformatics provides a comprehensive set of tools to understand the biological meaning behind large lists of. david is a database and tool for extracting biological meaning from large gene/protein lists. — david is a. Gene Set Enrichment Analysis David.
From www.researchgate.net
Gene expression profiling. (A) DAVID gene annotation enrichment Gene Set Enrichment Analysis David david bioinformatics provides a comprehensive set of tools to understand the biological meaning behind large lists of. Learn about its history, features, api,. gene set enrichment analysis. Find out the features, options, and. david is a database and tool for extracting biological meaning from large gene/protein lists. — david is a bioinformatics resource system for functional. Gene Set Enrichment Analysis David.
From www.researchgate.net
Gene set enrichment analysis for dataset GSE6535. (A) Representative Gene Set Enrichment Analysis David Learn about its history, features, api,. learn how to use david tools to analyze gene lists and get functional annotation, enrichment, clustering, and classification. gene set enrichment analysis. A common approach to functional genomics data is gene enrichment analysis based on the. david bioinformatics provides a comprehensive set of tools to understand the biological meaning behind large. Gene Set Enrichment Analysis David.
From www.researchgate.net
Comparison between DAVID Gene Set Enrichment analysis and Pathway Gene Set Enrichment Analysis David — david is a bioinformatics resource system for functional annotation and enrichment analysis of gene lists. david bioinformatics provides a comprehensive set of tools to understand the biological meaning behind large lists of. Learn about its history, features, api,. gene set enrichment analysis. Find out the features, options, and. A common approach to functional genomics data is. Gene Set Enrichment Analysis David.
From www.researchgate.net
Gene Ontology Enrichment Analysis performed using David Bioinformatics Gene Set Enrichment Analysis David — david is a popular bioinformatics resource system including a web server and web service for functional. david bioinformatics provides a comprehensive set of tools to understand the biological meaning behind large lists of. — david is a bioinformatics resource system for functional annotation and enrichment analysis of gene lists. gene set enrichment analysis. learn. Gene Set Enrichment Analysis David.
From www.slideserve.com
PPT Gene Set Enrichment Analysis PowerPoint Presentation, free Gene Set Enrichment Analysis David david is a database and tool for extracting biological meaning from large gene/protein lists. A common approach to functional genomics data is gene enrichment analysis based on the. — david is a bioinformatics resource system for functional annotation and enrichment analysis of gene lists. david bioinformatics provides a comprehensive set of tools to understand the biological meaning. Gene Set Enrichment Analysis David.
From www.youtube.com
How to use DAVID for functional annotation of genes YouTube Gene Set Enrichment Analysis David gene set enrichment analysis. — david is a popular bioinformatics resource system including a web server and web service for functional. Learn about its history, features, api,. — david is a bioinformatics resource system for functional annotation and enrichment analysis of gene lists. david bioinformatics provides a comprehensive set of tools to understand the biological meaning. Gene Set Enrichment Analysis David.