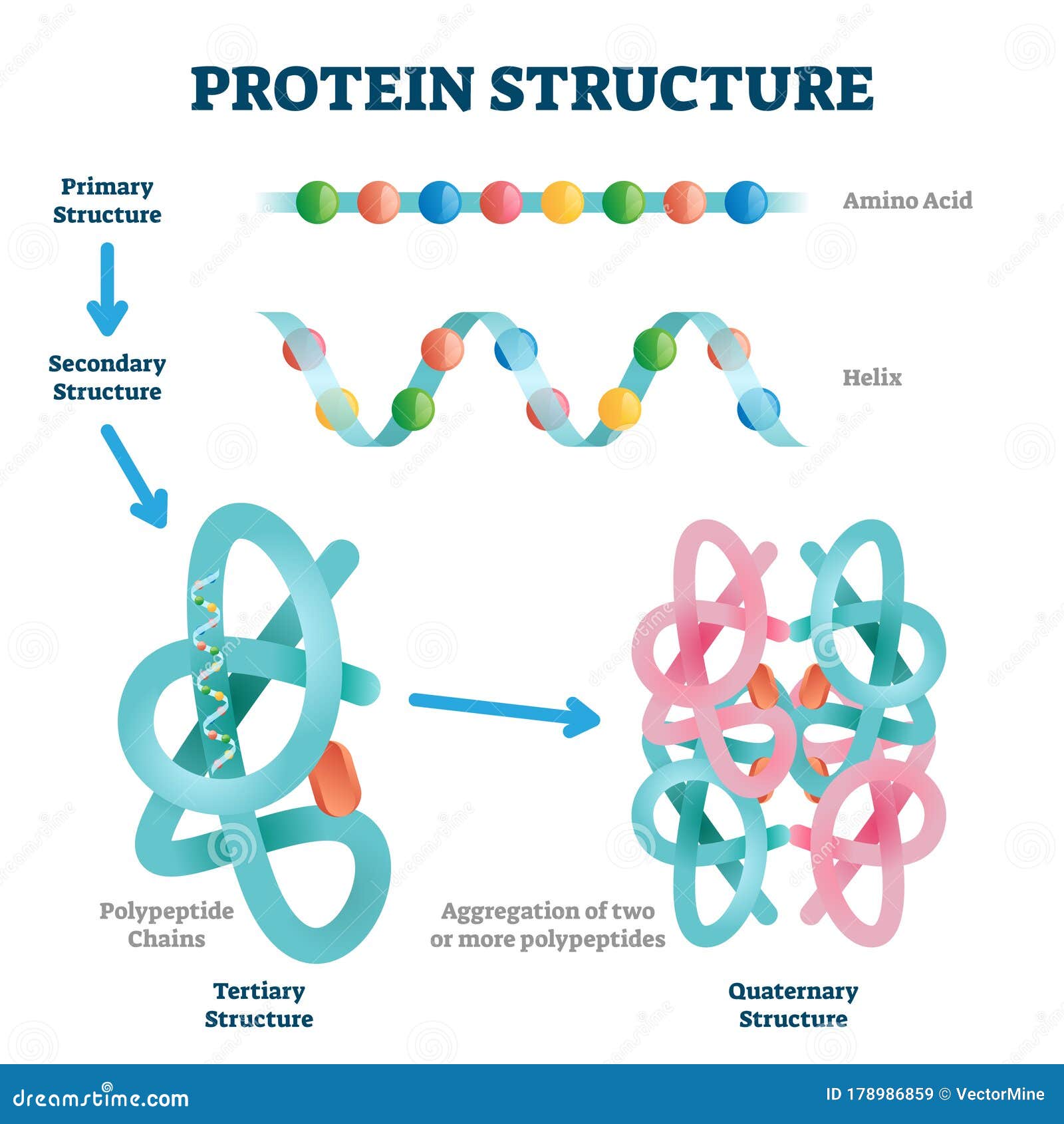
Amino Acids Structure Models . Download the simulation to start building proteins. Calculates and projects molecular electrostatic potential on a translucent or opaque van der waals surface. Every week we model all the sequences for thirteen core species based on the latest uniprotkb proteome. The rcsb pdb also provides a variety. There are 22 amino acids that are found in proteins and of these, only 20 are specified by the universal. This simulation allows students to combine amino acids to create proteins. Alphafold is an ai system developed by google deepmind that predicts a protein’s 3d structure from its amino acid sequence. As a member of the wwpdb, the rcsb pdb curates and annotates pdb data according to agreed upon standards.
from cartoondealer.com
As a member of the wwpdb, the rcsb pdb curates and annotates pdb data according to agreed upon standards. Alphafold is an ai system developed by google deepmind that predicts a protein’s 3d structure from its amino acid sequence. Every week we model all the sequences for thirteen core species based on the latest uniprotkb proteome. There are 22 amino acids that are found in proteins and of these, only 20 are specified by the universal. The rcsb pdb also provides a variety. This simulation allows students to combine amino acids to create proteins. Download the simulation to start building proteins. Calculates and projects molecular electrostatic potential on a translucent or opaque van der waals surface.
Amino Acid Tyrosine Molecular Structure RoyaltyFree Stock Image
Amino Acids Structure Models This simulation allows students to combine amino acids to create proteins. The rcsb pdb also provides a variety. Alphafold is an ai system developed by google deepmind that predicts a protein’s 3d structure from its amino acid sequence. This simulation allows students to combine amino acids to create proteins. Calculates and projects molecular electrostatic potential on a translucent or opaque van der waals surface. As a member of the wwpdb, the rcsb pdb curates and annotates pdb data according to agreed upon standards. There are 22 amino acids that are found in proteins and of these, only 20 are specified by the universal. Every week we model all the sequences for thirteen core species based on the latest uniprotkb proteome. Download the simulation to start building proteins.
From www.dreamstime.com
Amino acid stock vector. Illustration of nitrogen, nutrition 49550262 Amino Acids Structure Models This simulation allows students to combine amino acids to create proteins. As a member of the wwpdb, the rcsb pdb curates and annotates pdb data according to agreed upon standards. Alphafold is an ai system developed by google deepmind that predicts a protein’s 3d structure from its amino acid sequence. There are 22 amino acids that are found in proteins. Amino Acids Structure Models.
From www.shutterstock.com
Amino Acids Main Structural Chemical Formulas, 3d Illustration On Amino Acids Structure Models Alphafold is an ai system developed by google deepmind that predicts a protein’s 3d structure from its amino acid sequence. Every week we model all the sequences for thirteen core species based on the latest uniprotkb proteome. Download the simulation to start building proteins. The rcsb pdb also provides a variety. There are 22 amino acids that are found in. Amino Acids Structure Models.
From microbenotes.com
Amino Acids Properties, Structure, Classification, Functions Amino Acids Structure Models There are 22 amino acids that are found in proteins and of these, only 20 are specified by the universal. Calculates and projects molecular electrostatic potential on a translucent or opaque van der waals surface. Alphafold is an ai system developed by google deepmind that predicts a protein’s 3d structure from its amino acid sequence. The rcsb pdb also provides. Amino Acids Structure Models.
From www.dreamstime.com
Histidine. His C6H9N3O2. αAmino Acid. Structural Chemical Formula and Amino Acids Structure Models There are 22 amino acids that are found in proteins and of these, only 20 are specified by the universal. Calculates and projects molecular electrostatic potential on a translucent or opaque van der waals surface. The rcsb pdb also provides a variety. Alphafold is an ai system developed by google deepmind that predicts a protein’s 3d structure from its amino. Amino Acids Structure Models.
From www.expii.com
Amino Acids — Overview & Structure Expii Amino Acids Structure Models This simulation allows students to combine amino acids to create proteins. As a member of the wwpdb, the rcsb pdb curates and annotates pdb data according to agreed upon standards. Alphafold is an ai system developed by google deepmind that predicts a protein’s 3d structure from its amino acid sequence. There are 22 amino acids that are found in proteins. Amino Acids Structure Models.
From www.reagent.co.uk
What Are Amino Acids? The Science Blog Amino Acids Structure Models Alphafold is an ai system developed by google deepmind that predicts a protein’s 3d structure from its amino acid sequence. Download the simulation to start building proteins. As a member of the wwpdb, the rcsb pdb curates and annotates pdb data according to agreed upon standards. This simulation allows students to combine amino acids to create proteins. The rcsb pdb. Amino Acids Structure Models.
From www.thoughtco.com
Amino Acids Structure, Classification and Function Amino Acids Structure Models The rcsb pdb also provides a variety. Alphafold is an ai system developed by google deepmind that predicts a protein’s 3d structure from its amino acid sequence. There are 22 amino acids that are found in proteins and of these, only 20 are specified by the universal. Calculates and projects molecular electrostatic potential on a translucent or opaque van der. Amino Acids Structure Models.
From repone.de
Amino acids. 2D chemical structures of the 20 common amino acids REP ONE Amino Acids Structure Models Calculates and projects molecular electrostatic potential on a translucent or opaque van der waals surface. Every week we model all the sequences for thirteen core species based on the latest uniprotkb proteome. This simulation allows students to combine amino acids to create proteins. The rcsb pdb also provides a variety. Alphafold is an ai system developed by google deepmind that. Amino Acids Structure Models.
From chem.libretexts.org
13.1 Amino Acids Chemistry LibreTexts Amino Acids Structure Models This simulation allows students to combine amino acids to create proteins. As a member of the wwpdb, the rcsb pdb curates and annotates pdb data according to agreed upon standards. Alphafold is an ai system developed by google deepmind that predicts a protein’s 3d structure from its amino acid sequence. Calculates and projects molecular electrostatic potential on a translucent or. Amino Acids Structure Models.
From www.animalia-life.club
Amino Acid Model Amino Acids Structure Models Every week we model all the sequences for thirteen core species based on the latest uniprotkb proteome. This simulation allows students to combine amino acids to create proteins. There are 22 amino acids that are found in proteins and of these, only 20 are specified by the universal. Alphafold is an ai system developed by google deepmind that predicts a. Amino Acids Structure Models.
From www.alamy.com
Chemical formula, structural formula and 3D ballandstick model of L Amino Acids Structure Models There are 22 amino acids that are found in proteins and of these, only 20 are specified by the universal. This simulation allows students to combine amino acids to create proteins. Alphafold is an ai system developed by google deepmind that predicts a protein’s 3d structure from its amino acid sequence. Download the simulation to start building proteins. Calculates and. Amino Acids Structure Models.
From www.thoughtco.com
Amino Acids Structure, Classification and Function Amino Acids Structure Models Alphafold is an ai system developed by google deepmind that predicts a protein’s 3d structure from its amino acid sequence. Calculates and projects molecular electrostatic potential on a translucent or opaque van der waals surface. This simulation allows students to combine amino acids to create proteins. The rcsb pdb also provides a variety. Download the simulation to start building proteins.. Amino Acids Structure Models.
From www.ib.bioninja.com.au
Amino Acids Amino Acids Structure Models Download the simulation to start building proteins. This simulation allows students to combine amino acids to create proteins. The rcsb pdb also provides a variety. As a member of the wwpdb, the rcsb pdb curates and annotates pdb data according to agreed upon standards. Calculates and projects molecular electrostatic potential on a translucent or opaque van der waals surface. Alphafold. Amino Acids Structure Models.
From klasyrkhl.blob.core.windows.net
Amino Acid Chain Structure at Mark Ritter blog Amino Acids Structure Models Alphafold is an ai system developed by google deepmind that predicts a protein’s 3d structure from its amino acid sequence. The rcsb pdb also provides a variety. There are 22 amino acids that are found in proteins and of these, only 20 are specified by the universal. This simulation allows students to combine amino acids to create proteins. As a. Amino Acids Structure Models.
From www.alamy.com
Protein structure. Amino acids, peptides, protein. Proteins formation Amino Acids Structure Models Download the simulation to start building proteins. Every week we model all the sequences for thirteen core species based on the latest uniprotkb proteome. There are 22 amino acids that are found in proteins and of these, only 20 are specified by the universal. This simulation allows students to combine amino acids to create proteins. Calculates and projects molecular electrostatic. Amino Acids Structure Models.
From www.indigoinstruments.com
Tryptophan Structure Amino Acid Molecular Model, Molymod Style Amino Acids Structure Models Alphafold is an ai system developed by google deepmind that predicts a protein’s 3d structure from its amino acid sequence. There are 22 amino acids that are found in proteins and of these, only 20 are specified by the universal. As a member of the wwpdb, the rcsb pdb curates and annotates pdb data according to agreed upon standards. The. Amino Acids Structure Models.
From www.britannica.com
Video of amino acids Britannica Amino Acids Structure Models The rcsb pdb also provides a variety. Every week we model all the sequences for thirteen core species based on the latest uniprotkb proteome. There are 22 amino acids that are found in proteins and of these, only 20 are specified by the universal. This simulation allows students to combine amino acids to create proteins. As a member of the. Amino Acids Structure Models.
From www.creative-biostructure.com
Amino Acid Properties and Structure Creative Biostructure Amino Acids Structure Models Alphafold is an ai system developed by google deepmind that predicts a protein’s 3d structure from its amino acid sequence. There are 22 amino acids that are found in proteins and of these, only 20 are specified by the universal. As a member of the wwpdb, the rcsb pdb curates and annotates pdb data according to agreed upon standards. This. Amino Acids Structure Models.
From www.mun.ca
Amino Acid Structure Amino Acids Structure Models There are 22 amino acids that are found in proteins and of these, only 20 are specified by the universal. This simulation allows students to combine amino acids to create proteins. Calculates and projects molecular electrostatic potential on a translucent or opaque van der waals surface. Every week we model all the sequences for thirteen core species based on the. Amino Acids Structure Models.
From www.alamy.com
20 Amino Acid Molecules Ball and Stick Structure Stock Vector Image Amino Acids Structure Models Download the simulation to start building proteins. This simulation allows students to combine amino acids to create proteins. Every week we model all the sequences for thirteen core species based on the latest uniprotkb proteome. There are 22 amino acids that are found in proteins and of these, only 20 are specified by the universal. The rcsb pdb also provides. Amino Acids Structure Models.
From www.alamy.com
General formula of amino acids, ionized and nonionized (zwitterion Amino Acids Structure Models There are 22 amino acids that are found in proteins and of these, only 20 are specified by the universal. Calculates and projects molecular electrostatic potential on a translucent or opaque van der waals surface. This simulation allows students to combine amino acids to create proteins. Every week we model all the sequences for thirteen core species based on the. Amino Acids Structure Models.
From courses.lumenlearning.com
Amino Acids Structure Nutrition Amino Acids Structure Models Download the simulation to start building proteins. Calculates and projects molecular electrostatic potential on a translucent or opaque van der waals surface. There are 22 amino acids that are found in proteins and of these, only 20 are specified by the universal. This simulation allows students to combine amino acids to create proteins. As a member of the wwpdb, the. Amino Acids Structure Models.
From www.creative-biostructure.com
Amino Acid Properties and Structure Creative Biostructure Amino Acids Structure Models As a member of the wwpdb, the rcsb pdb curates and annotates pdb data according to agreed upon standards. Every week we model all the sequences for thirteen core species based on the latest uniprotkb proteome. Calculates and projects molecular electrostatic potential on a translucent or opaque van der waals surface. This simulation allows students to combine amino acids to. Amino Acids Structure Models.
From sketchfab.com
Amino Acid Structure 3D model by Andy Todd (atodd19) [53a46b8 Amino Acids Structure Models Alphafold is an ai system developed by google deepmind that predicts a protein’s 3d structure from its amino acid sequence. Every week we model all the sequences for thirteen core species based on the latest uniprotkb proteome. There are 22 amino acids that are found in proteins and of these, only 20 are specified by the universal. This simulation allows. Amino Acids Structure Models.
From stock.adobe.com
Vektorová grafika „illustration of chemistry, Protein structure model Amino Acids Structure Models Calculates and projects molecular electrostatic potential on a translucent or opaque van der waals surface. This simulation allows students to combine amino acids to create proteins. Download the simulation to start building proteins. There are 22 amino acids that are found in proteins and of these, only 20 are specified by the universal. Every week we model all the sequences. Amino Acids Structure Models.
From www.alamy.com
Chemical formula, structural formula and 3D ballandstick model of Amino Acids Structure Models Alphafold is an ai system developed by google deepmind that predicts a protein’s 3d structure from its amino acid sequence. The rcsb pdb also provides a variety. There are 22 amino acids that are found in proteins and of these, only 20 are specified by the universal. Every week we model all the sequences for thirteen core species based on. Amino Acids Structure Models.
From www.dreamstime.com
Amino Acid. Structural Formula and Model of Molecule Stock Vector Amino Acids Structure Models Every week we model all the sequences for thirteen core species based on the latest uniprotkb proteome. Alphafold is an ai system developed by google deepmind that predicts a protein’s 3d structure from its amino acid sequence. As a member of the wwpdb, the rcsb pdb curates and annotates pdb data according to agreed upon standards. There are 22 amino. Amino Acids Structure Models.
From cartoondealer.com
Amino Acid Tyrosine Molecular Structure RoyaltyFree Stock Image Amino Acids Structure Models Alphafold is an ai system developed by google deepmind that predicts a protein’s 3d structure from its amino acid sequence. The rcsb pdb also provides a variety. Calculates and projects molecular electrostatic potential on a translucent or opaque van der waals surface. Download the simulation to start building proteins. As a member of the wwpdb, the rcsb pdb curates and. Amino Acids Structure Models.
From www.dreamstime.com
Amino Acid Labeled Diagram Vector Illustration Drawing Biochemistry Amino Acids Structure Models Download the simulation to start building proteins. There are 22 amino acids that are found in proteins and of these, only 20 are specified by the universal. Every week we model all the sequences for thirteen core species based on the latest uniprotkb proteome. This simulation allows students to combine amino acids to create proteins. As a member of the. Amino Acids Structure Models.
From www.alamy.com
General formula of amino acids, which are building blocks of proteins Amino Acids Structure Models Every week we model all the sequences for thirteen core species based on the latest uniprotkb proteome. Alphafold is an ai system developed by google deepmind that predicts a protein’s 3d structure from its amino acid sequence. This simulation allows students to combine amino acids to create proteins. Download the simulation to start building proteins. There are 22 amino acids. Amino Acids Structure Models.
From www.turbosquid.com
amino acids 3d obj Amino Acids Structure Models As a member of the wwpdb, the rcsb pdb curates and annotates pdb data according to agreed upon standards. This simulation allows students to combine amino acids to create proteins. There are 22 amino acids that are found in proteins and of these, only 20 are specified by the universal. The rcsb pdb also provides a variety. Every week we. Amino Acids Structure Models.
From mungfali.com
Amino Acid Structure Chart Amino Acids Structure Models This simulation allows students to combine amino acids to create proteins. Download the simulation to start building proteins. Alphafold is an ai system developed by google deepmind that predicts a protein’s 3d structure from its amino acid sequence. Calculates and projects molecular electrostatic potential on a translucent or opaque van der waals surface. As a member of the wwpdb, the. Amino Acids Structure Models.
From www.alamy.com
Protein. Structural chemical formula and molecular model. General Amino Acids Structure Models The rcsb pdb also provides a variety. Calculates and projects molecular electrostatic potential on a translucent or opaque van der waals surface. Every week we model all the sequences for thirteen core species based on the latest uniprotkb proteome. As a member of the wwpdb, the rcsb pdb curates and annotates pdb data according to agreed upon standards. There are. Amino Acids Structure Models.
From www.animalia-life.club
Basic Amino Acid Structure Amino Acids Structure Models There are 22 amino acids that are found in proteins and of these, only 20 are specified by the universal. Download the simulation to start building proteins. The rcsb pdb also provides a variety. Every week we model all the sequences for thirteen core species based on the latest uniprotkb proteome. This simulation allows students to combine amino acids to. Amino Acids Structure Models.
From www.compoundchem.com
A Brief Guide to the Twenty Common Amino Acids Compound Interest Amino Acids Structure Models This simulation allows students to combine amino acids to create proteins. Every week we model all the sequences for thirteen core species based on the latest uniprotkb proteome. Download the simulation to start building proteins. Calculates and projects molecular electrostatic potential on a translucent or opaque van der waals surface. As a member of the wwpdb, the rcsb pdb curates. Amino Acids Structure Models.