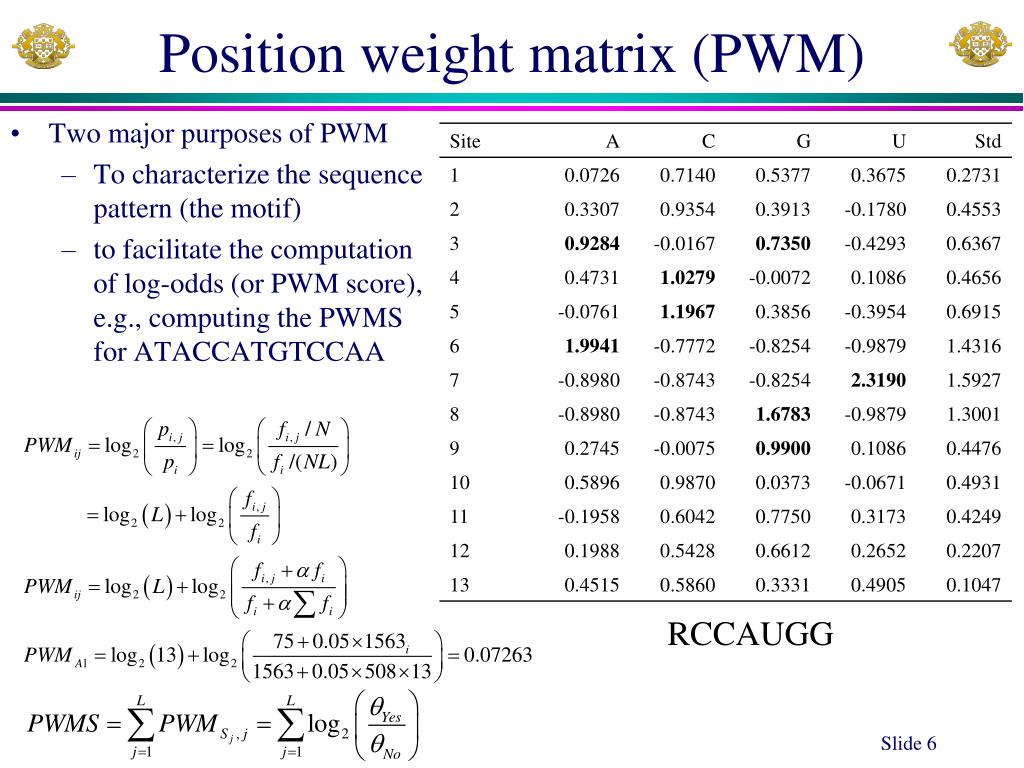
Position Weight Matrix . position weight matrix (pwm) is a scoring matrix composed of the log likelihood of each nucleotide in a motif (tfbs). learn how to construct and use position weight matrices (pwm) to model transcription factor binding sites (tfbs) in biological sequences. learn how to use position weight matrices (pwms) to describe and scan short sequences, such as transcription factor. learn about position weight matrix (pwm), a scoring matrix composed of the log likelihood of each nucleotide in a motif. See examples, formulae, pseudocount function and r code. position weight matrix (pwm) is not only one of the most widely used.
from www.slideserve.com
learn about position weight matrix (pwm), a scoring matrix composed of the log likelihood of each nucleotide in a motif. learn how to use position weight matrices (pwms) to describe and scan short sequences, such as transcription factor. position weight matrix (pwm) is not only one of the most widely used. See examples, formulae, pseudocount function and r code. position weight matrix (pwm) is a scoring matrix composed of the log likelihood of each nucleotide in a motif (tfbs). learn how to construct and use position weight matrices (pwm) to model transcription factor binding sites (tfbs) in biological sequences.
PPT Position weight matrix (PWM), Perceptron and their applications
Position Weight Matrix position weight matrix (pwm) is a scoring matrix composed of the log likelihood of each nucleotide in a motif (tfbs). learn how to construct and use position weight matrices (pwm) to model transcription factor binding sites (tfbs) in biological sequences. learn how to use position weight matrices (pwms) to describe and scan short sequences, such as transcription factor. See examples, formulae, pseudocount function and r code. position weight matrix (pwm) is not only one of the most widely used. position weight matrix (pwm) is a scoring matrix composed of the log likelihood of each nucleotide in a motif (tfbs). learn about position weight matrix (pwm), a scoring matrix composed of the log likelihood of each nucleotide in a motif.
From www.researchgate.net
Weight matrices and sequence encoding. (A) The weight matrix for a Position Weight Matrix learn about position weight matrix (pwm), a scoring matrix composed of the log likelihood of each nucleotide in a motif. learn how to use position weight matrices (pwms) to describe and scan short sequences, such as transcription factor. See examples, formulae, pseudocount function and r code. position weight matrix (pwm) is a scoring matrix composed of the. Position Weight Matrix.
From www.slideserve.com
PPT DNA Motif Finding PowerPoint Presentation, free download ID1134511 Position Weight Matrix learn how to use position weight matrices (pwms) to describe and scan short sequences, such as transcription factor. position weight matrix (pwm) is a scoring matrix composed of the log likelihood of each nucleotide in a motif (tfbs). learn about position weight matrix (pwm), a scoring matrix composed of the log likelihood of each nucleotide in a. Position Weight Matrix.
From dxoaciugb.blob.core.windows.net
Position Weight Matrix In R at Timothy Isakson blog Position Weight Matrix learn how to use position weight matrices (pwms) to describe and scan short sequences, such as transcription factor. position weight matrix (pwm) is a scoring matrix composed of the log likelihood of each nucleotide in a motif (tfbs). See examples, formulae, pseudocount function and r code. position weight matrix (pwm) is not only one of the most. Position Weight Matrix.
From www.researchgate.net
Models for proteinDNA binding preferences. (A) Position weight matrix Position Weight Matrix position weight matrix (pwm) is not only one of the most widely used. learn how to use position weight matrices (pwms) to describe and scan short sequences, such as transcription factor. See examples, formulae, pseudocount function and r code. learn how to construct and use position weight matrices (pwm) to model transcription factor binding sites (tfbs) in. Position Weight Matrix.
From www.researchgate.net
Position weight matrices depicting binding specificities of Tbr Position Weight Matrix learn how to construct and use position weight matrices (pwm) to model transcription factor binding sites (tfbs) in biological sequences. learn how to use position weight matrices (pwms) to describe and scan short sequences, such as transcription factor. position weight matrix (pwm) is a scoring matrix composed of the log likelihood of each nucleotide in a motif. Position Weight Matrix.
From www.researchgate.net
Position weight matrices depicting binding specificities of Tbr Position Weight Matrix See examples, formulae, pseudocount function and r code. position weight matrix (pwm) is not only one of the most widely used. learn about position weight matrix (pwm), a scoring matrix composed of the log likelihood of each nucleotide in a motif. position weight matrix (pwm) is a scoring matrix composed of the log likelihood of each nucleotide. Position Weight Matrix.
From www.slideserve.com
PPT DNA Regulatory Binding Motif Search PowerPoint Presentation, free Position Weight Matrix position weight matrix (pwm) is a scoring matrix composed of the log likelihood of each nucleotide in a motif (tfbs). See examples, formulae, pseudocount function and r code. learn how to construct and use position weight matrices (pwm) to model transcription factor binding sites (tfbs) in biological sequences. learn about position weight matrix (pwm), a scoring matrix. Position Weight Matrix.
From www.researchgate.net
The number of unique transcription factors with position weight Position Weight Matrix learn how to construct and use position weight matrices (pwm) to model transcription factor binding sites (tfbs) in biological sequences. See examples, formulae, pseudocount function and r code. learn how to use position weight matrices (pwms) to describe and scan short sequences, such as transcription factor. position weight matrix (pwm) is not only one of the most. Position Weight Matrix.
From www.slideserve.com
PPT Gene prediction PowerPoint Presentation, free download ID4237759 Position Weight Matrix learn how to construct and use position weight matrices (pwm) to model transcription factor binding sites (tfbs) in biological sequences. learn about position weight matrix (pwm), a scoring matrix composed of the log likelihood of each nucleotide in a motif. position weight matrix (pwm) is not only one of the most widely used. position weight matrix. Position Weight Matrix.
From www.researchgate.net
1 Creating a Position Weight Matrix (PWM). The nucleotide counts at Position Weight Matrix learn about position weight matrix (pwm), a scoring matrix composed of the log likelihood of each nucleotide in a motif. learn how to use position weight matrices (pwms) to describe and scan short sequences, such as transcription factor. position weight matrix (pwm) is not only one of the most widely used. position weight matrix (pwm) is. Position Weight Matrix.
From www.slideserve.com
PPT Gene Structure and Identification III PowerPoint Presentation Position Weight Matrix position weight matrix (pwm) is a scoring matrix composed of the log likelihood of each nucleotide in a motif (tfbs). learn how to construct and use position weight matrices (pwm) to model transcription factor binding sites (tfbs) in biological sequences. learn how to use position weight matrices (pwms) to describe and scan short sequences, such as transcription. Position Weight Matrix.
From www.researchgate.net
Position weight matrices and mutation maps provide information about Position Weight Matrix position weight matrix (pwm) is a scoring matrix composed of the log likelihood of each nucleotide in a motif (tfbs). learn about position weight matrix (pwm), a scoring matrix composed of the log likelihood of each nucleotide in a motif. See examples, formulae, pseudocount function and r code. position weight matrix (pwm) is not only one of. Position Weight Matrix.
From www.slideserve.com
PPT Position weight matrix (PWM), Perceptron and their applications Position Weight Matrix learn how to use position weight matrices (pwms) to describe and scan short sequences, such as transcription factor. position weight matrix (pwm) is not only one of the most widely used. learn how to construct and use position weight matrices (pwm) to model transcription factor binding sites (tfbs) in biological sequences. learn about position weight matrix. Position Weight Matrix.
From www.slideserve.com
PPT Position weight matrix (PWM), Perceptron and their applications Position Weight Matrix position weight matrix (pwm) is a scoring matrix composed of the log likelihood of each nucleotide in a motif (tfbs). See examples, formulae, pseudocount function and r code. learn how to construct and use position weight matrices (pwm) to model transcription factor binding sites (tfbs) in biological sequences. position weight matrix (pwm) is not only one of. Position Weight Matrix.
From www.slideserve.com
PPT Position weight matrix (PWM), Perceptron and their applications Position Weight Matrix position weight matrix (pwm) is a scoring matrix composed of the log likelihood of each nucleotide in a motif (tfbs). See examples, formulae, pseudocount function and r code. position weight matrix (pwm) is not only one of the most widely used. learn about position weight matrix (pwm), a scoring matrix composed of the log likelihood of each. Position Weight Matrix.
From www.researchgate.net
Position frequency matrix (PFM), position probability matrix (PPM Position Weight Matrix See examples, formulae, pseudocount function and r code. position weight matrix (pwm) is a scoring matrix composed of the log likelihood of each nucleotide in a motif (tfbs). learn how to use position weight matrices (pwms) to describe and scan short sequences, such as transcription factor. learn how to construct and use position weight matrices (pwm) to. Position Weight Matrix.
From www.slideserve.com
PPT Counting position weight matrices in a sequence & an application Position Weight Matrix learn about position weight matrix (pwm), a scoring matrix composed of the log likelihood of each nucleotide in a motif. See examples, formulae, pseudocount function and r code. position weight matrix (pwm) is not only one of the most widely used. learn how to use position weight matrices (pwms) to describe and scan short sequences, such as. Position Weight Matrix.
From www.researchgate.net
Establishing an MeCP2 position weight matrix threshold and GC Position Weight Matrix See examples, formulae, pseudocount function and r code. learn how to construct and use position weight matrices (pwm) to model transcription factor binding sites (tfbs) in biological sequences. position weight matrix (pwm) is a scoring matrix composed of the log likelihood of each nucleotide in a motif (tfbs). learn how to use position weight matrices (pwms) to. Position Weight Matrix.
From www.researchgate.net
(PDF) TreeBased Position Weight Matrix Approach to Model Transcription Position Weight Matrix learn how to construct and use position weight matrices (pwm) to model transcription factor binding sites (tfbs) in biological sequences. learn how to use position weight matrices (pwms) to describe and scan short sequences, such as transcription factor. position weight matrix (pwm) is a scoring matrix composed of the log likelihood of each nucleotide in a motif. Position Weight Matrix.
From www.chegg.com
Solved Position Weight Matrices Below is what is known as a Position Weight Matrix learn about position weight matrix (pwm), a scoring matrix composed of the log likelihood of each nucleotide in a motif. learn how to use position weight matrices (pwms) to describe and scan short sequences, such as transcription factor. position weight matrix (pwm) is not only one of the most widely used. position weight matrix (pwm) is. Position Weight Matrix.
From www.researchgate.net
Position weight matrices and mutation maps provide information about Position Weight Matrix learn how to construct and use position weight matrices (pwm) to model transcription factor binding sites (tfbs) in biological sequences. learn how to use position weight matrices (pwms) to describe and scan short sequences, such as transcription factor. learn about position weight matrix (pwm), a scoring matrix composed of the log likelihood of each nucleotide in a. Position Weight Matrix.
From www.researchgate.net
Models for proteinDNA binding preferences. (A) Position weight matrix Position Weight Matrix See examples, formulae, pseudocount function and r code. position weight matrix (pwm) is not only one of the most widely used. learn about position weight matrix (pwm), a scoring matrix composed of the log likelihood of each nucleotide in a motif. learn how to use position weight matrices (pwms) to describe and scan short sequences, such as. Position Weight Matrix.
From davetang.org
Position weight matrix Dave Tang's blog Position Weight Matrix learn how to use position weight matrices (pwms) to describe and scan short sequences, such as transcription factor. learn how to construct and use position weight matrices (pwm) to model transcription factor binding sites (tfbs) in biological sequences. See examples, formulae, pseudocount function and r code. learn about position weight matrix (pwm), a scoring matrix composed of. Position Weight Matrix.
From www.researchgate.net
Position weight matrix view of PRODORIC for the binding site of the Anr Position Weight Matrix learn about position weight matrix (pwm), a scoring matrix composed of the log likelihood of each nucleotide in a motif. learn how to construct and use position weight matrices (pwm) to model transcription factor binding sites (tfbs) in biological sequences. learn how to use position weight matrices (pwms) to describe and scan short sequences, such as transcription. Position Weight Matrix.
From www.slideserve.com
PPT Position weight matrix (PWM), Perceptron and their applications Position Weight Matrix learn how to construct and use position weight matrices (pwm) to model transcription factor binding sites (tfbs) in biological sequences. position weight matrix (pwm) is not only one of the most widely used. position weight matrix (pwm) is a scoring matrix composed of the log likelihood of each nucleotide in a motif (tfbs). See examples, formulae, pseudocount. Position Weight Matrix.
From www.researchgate.net
DNAbinding specificity of plant TFs. Position weight matrix (PWM Position Weight Matrix learn how to construct and use position weight matrices (pwm) to model transcription factor binding sites (tfbs) in biological sequences. position weight matrix (pwm) is a scoring matrix composed of the log likelihood of each nucleotide in a motif (tfbs). See examples, formulae, pseudocount function and r code. learn how to use position weight matrices (pwms) to. Position Weight Matrix.
From www.slideserve.com
PPT CS5263 Bioinformatics PowerPoint Presentation, free download ID Position Weight Matrix position weight matrix (pwm) is not only one of the most widely used. See examples, formulae, pseudocount function and r code. learn how to construct and use position weight matrices (pwm) to model transcription factor binding sites (tfbs) in biological sequences. learn how to use position weight matrices (pwms) to describe and scan short sequences, such as. Position Weight Matrix.
From www.researchgate.net
Position Weight Matrices and Sequence Alignments of Selected Position Weight Matrix position weight matrix (pwm) is a scoring matrix composed of the log likelihood of each nucleotide in a motif (tfbs). learn how to construct and use position weight matrices (pwm) to model transcription factor binding sites (tfbs) in biological sequences. See examples, formulae, pseudocount function and r code. learn about position weight matrix (pwm), a scoring matrix. Position Weight Matrix.
From www.slideserve.com
PPT Transcription factor binding sites and gene regulatory network Position Weight Matrix learn how to construct and use position weight matrices (pwm) to model transcription factor binding sites (tfbs) in biological sequences. learn how to use position weight matrices (pwms) to describe and scan short sequences, such as transcription factor. position weight matrix (pwm) is a scoring matrix composed of the log likelihood of each nucleotide in a motif. Position Weight Matrix.
From www.slideserve.com
PPT Position weight matrix (PWM), Perceptron and their applications Position Weight Matrix See examples, formulae, pseudocount function and r code. learn how to construct and use position weight matrices (pwm) to model transcription factor binding sites (tfbs) in biological sequences. position weight matrix (pwm) is a scoring matrix composed of the log likelihood of each nucleotide in a motif (tfbs). learn about position weight matrix (pwm), a scoring matrix. Position Weight Matrix.
From www.slideserve.com
PPT A System Approach to Measuring the Binding Energy Landscapes of Position Weight Matrix position weight matrix (pwm) is a scoring matrix composed of the log likelihood of each nucleotide in a motif (tfbs). position weight matrix (pwm) is not only one of the most widely used. See examples, formulae, pseudocount function and r code. learn about position weight matrix (pwm), a scoring matrix composed of the log likelihood of each. Position Weight Matrix.
From www.slideserve.com
PPT Position Weight Matrices for Representing Signals in Sequences Position Weight Matrix position weight matrix (pwm) is not only one of the most widely used. position weight matrix (pwm) is a scoring matrix composed of the log likelihood of each nucleotide in a motif (tfbs). learn about position weight matrix (pwm), a scoring matrix composed of the log likelihood of each nucleotide in a motif. learn how to. Position Weight Matrix.
From www.researchgate.net
Spearman correlation coefficients ( ) for position weight matrix (PWM Position Weight Matrix learn how to use position weight matrices (pwms) to describe and scan short sequences, such as transcription factor. position weight matrix (pwm) is not only one of the most widely used. learn how to construct and use position weight matrices (pwm) to model transcription factor binding sites (tfbs) in biological sequences. learn about position weight matrix. Position Weight Matrix.
From www.researchgate.net
2 Position weight matrix and motif scoring (A) Align known binding Position Weight Matrix learn how to use position weight matrices (pwms) to describe and scan short sequences, such as transcription factor. See examples, formulae, pseudocount function and r code. learn how to construct and use position weight matrices (pwm) to model transcription factor binding sites (tfbs) in biological sequences. position weight matrix (pwm) is not only one of the most. Position Weight Matrix.
From www.slideserve.com
PPT Position weight matrix (PWM), Perceptron and their applications Position Weight Matrix learn about position weight matrix (pwm), a scoring matrix composed of the log likelihood of each nucleotide in a motif. position weight matrix (pwm) is not only one of the most widely used. See examples, formulae, pseudocount function and r code. learn how to construct and use position weight matrices (pwm) to model transcription factor binding sites. Position Weight Matrix.