Blast Protein Comparison . This tutorial will explain how to compare and identify variation between two or more protein sequences. A new approach to rapid sequence comparison, basic local alignment search tool (blast), directly approximates alignments that optimize a measure of local similarity, the maximal. The basic local alignment search tool (blast) finds regions of local similarity between protein or nucleotide sequences. Compares one or more protein query sequences to a subject protein sequence or a database of protein sequences. The program compares nucleotide or protein. There are versions of blast. This is useful when trying to. The basic local alignment search tool (blast) finds regions of local similarity between sequences. Blastp simply compares a protein query to a protein database. The program compares nucleotide or protein sequences to sequence. The basic local alignment search tool (blast) is the most widely used sequence similarity tool. See also, the blast tutorial series:.
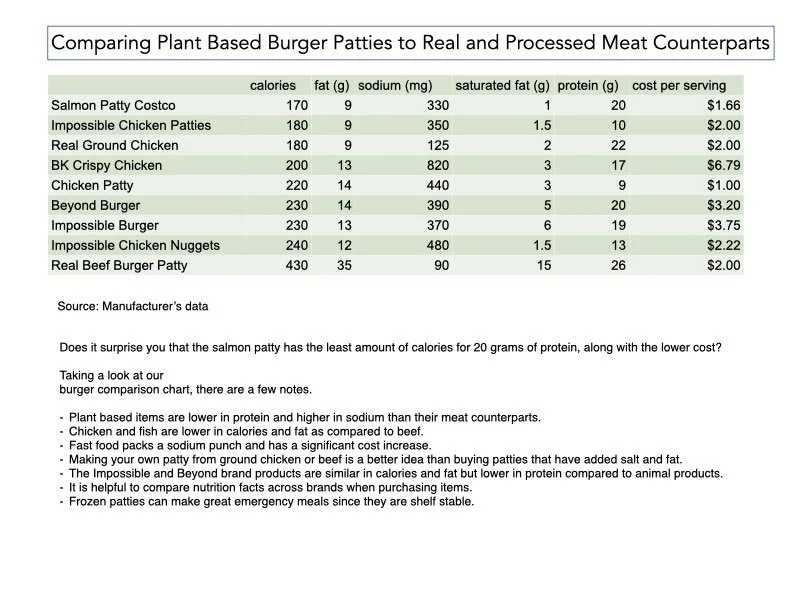
from www.foodandhealth.com
A new approach to rapid sequence comparison, basic local alignment search tool (blast), directly approximates alignments that optimize a measure of local similarity, the maximal. The basic local alignment search tool (blast) is the most widely used sequence similarity tool. The program compares nucleotide or protein. Compares one or more protein query sequences to a subject protein sequence or a database of protein sequences. See also, the blast tutorial series:. There are versions of blast. The program compares nucleotide or protein sequences to sequence. This tutorial will explain how to compare and identify variation between two or more protein sequences. Blastp simply compares a protein query to a protein database. This is useful when trying to.
Protein Comparison PDF Printable Handout — Food and Health Communications
Blast Protein Comparison Blastp simply compares a protein query to a protein database. A new approach to rapid sequence comparison, basic local alignment search tool (blast), directly approximates alignments that optimize a measure of local similarity, the maximal. The program compares nucleotide or protein. The program compares nucleotide or protein sequences to sequence. There are versions of blast. Compares one or more protein query sequences to a subject protein sequence or a database of protein sequences. This tutorial will explain how to compare and identify variation between two or more protein sequences. This is useful when trying to. The basic local alignment search tool (blast) is the most widely used sequence similarity tool. The basic local alignment search tool (blast) finds regions of local similarity between protein or nucleotide sequences. The basic local alignment search tool (blast) finds regions of local similarity between sequences. Blastp simply compares a protein query to a protein database. See also, the blast tutorial series:.
From dokumen.tips
(PDF) 3DBLAST 3D PROTEIN STRUCTURE ALIGNMENT, COMPARISON, …psb Blast Protein Comparison A new approach to rapid sequence comparison, basic local alignment search tool (blast), directly approximates alignments that optimize a measure of local similarity, the maximal. The basic local alignment search tool (blast) finds regions of local similarity between protein or nucleotide sequences. Compares one or more protein query sequences to a subject protein sequence or a database of protein sequences.. Blast Protein Comparison.
From www.psb.ugent.be
PTM Viewer Blast Protein Comparison The basic local alignment search tool (blast) finds regions of local similarity between protein or nucleotide sequences. A new approach to rapid sequence comparison, basic local alignment search tool (blast), directly approximates alignments that optimize a measure of local similarity, the maximal. The program compares nucleotide or protein. This is useful when trying to. See also, the blast tutorial series:.. Blast Protein Comparison.
From slidetodoc.com
Bioinformatics 91 05 Sequence Comparison DNA vs Protein Blast Protein Comparison This tutorial will explain how to compare and identify variation between two or more protein sequences. Compares one or more protein query sequences to a subject protein sequence or a database of protein sequences. There are versions of blast. See also, the blast tutorial series:. Blastp simply compares a protein query to a protein database. This is useful when trying. Blast Protein Comparison.
From www.youtube.com
Translated Nucleotide to Protein BLAST BLASTX YouTube Blast Protein Comparison The basic local alignment search tool (blast) is the most widely used sequence similarity tool. There are versions of blast. Blastp simply compares a protein query to a protein database. See also, the blast tutorial series:. This is useful when trying to. Compares one or more protein query sequences to a subject protein sequence or a database of protein sequences.. Blast Protein Comparison.
From www.semanticscholar.org
Figure 1 from The BLAST Sequence Analysis Tool Semantic Scholar Blast Protein Comparison The program compares nucleotide or protein. The basic local alignment search tool (blast) finds regions of local similarity between sequences. A new approach to rapid sequence comparison, basic local alignment search tool (blast), directly approximates alignments that optimize a measure of local similarity, the maximal. The program compares nucleotide or protein sequences to sequence. The basic local alignment search tool. Blast Protein Comparison.
From abacus.bates.edu
BLASTP Results Part 1 Blast Protein Comparison This is useful when trying to. The basic local alignment search tool (blast) finds regions of local similarity between protein or nucleotide sequences. The program compares nucleotide or protein. The basic local alignment search tool (blast) finds regions of local similarity between sequences. There are versions of blast. This tutorial will explain how to compare and identify variation between two. Blast Protein Comparison.
From www.researchgate.net
List of NCBI database protein accession numbers used in the BLASTp Blast Protein Comparison Compares one or more protein query sequences to a subject protein sequence or a database of protein sequences. The program compares nucleotide or protein sequences to sequence. The basic local alignment search tool (blast) finds regions of local similarity between protein or nucleotide sequences. This is useful when trying to. The program compares nucleotide or protein. A new approach to. Blast Protein Comparison.
From dokumen.tips
(PDF) Introduction to BLAST with Protein Sequencesmath.usu.edu Blast Protein Comparison The basic local alignment search tool (blast) finds regions of local similarity between sequences. This tutorial will explain how to compare and identify variation between two or more protein sequences. The basic local alignment search tool (blast) is the most widely used sequence similarity tool. There are versions of blast. The basic local alignment search tool (blast) finds regions of. Blast Protein Comparison.
From www.researchgate.net
Scaling comparison of HSPBLAST and NCBI BLAST on Kraken. Weak scaling Blast Protein Comparison See also, the blast tutorial series:. This tutorial will explain how to compare and identify variation between two or more protein sequences. Blastp simply compares a protein query to a protein database. The basic local alignment search tool (blast) finds regions of local similarity between protein or nucleotide sequences. The program compares nucleotide or protein. The basic local alignment search. Blast Protein Comparison.
From www.slideserve.com
PPT Gapped BLAST and PSIBLAST a new generation of protein database Blast Protein Comparison The program compares nucleotide or protein sequences to sequence. There are versions of blast. Compares one or more protein query sequences to a subject protein sequence or a database of protein sequences. The basic local alignment search tool (blast) finds regions of local similarity between protein or nucleotide sequences. Blastp simply compares a protein query to a protein database. See. Blast Protein Comparison.
From www.slideserve.com
PPT Protein Sequence PowerPoint Presentation, free download ID4271810 Blast Protein Comparison The program compares nucleotide or protein. The basic local alignment search tool (blast) is the most widely used sequence similarity tool. A new approach to rapid sequence comparison, basic local alignment search tool (blast), directly approximates alignments that optimize a measure of local similarity, the maximal. The program compares nucleotide or protein sequences to sequence. This is useful when trying. Blast Protein Comparison.
From www.foodandhealth.com
Plant Protein Comparison Chart — Food and Health Communications Blast Protein Comparison The basic local alignment search tool (blast) finds regions of local similarity between protein or nucleotide sequences. Blastp simply compares a protein query to a protein database. The program compares nucleotide or protein. This tutorial will explain how to compare and identify variation between two or more protein sequences. The program compares nucleotide or protein sequences to sequence. A new. Blast Protein Comparison.
From pediaa.com
Difference Between BLAST and FASTA Definition, Features, Uses Blast Protein Comparison Compares one or more protein query sequences to a subject protein sequence or a database of protein sequences. The program compares nucleotide or protein sequences to sequence. A new approach to rapid sequence comparison, basic local alignment search tool (blast), directly approximates alignments that optimize a measure of local similarity, the maximal. See also, the blast tutorial series:. This is. Blast Protein Comparison.
From www.youtube.com
How To Analyze DNA Sequence Using BLAST Basic Local Alignment Search Blast Protein Comparison There are versions of blast. The basic local alignment search tool (blast) finds regions of local similarity between protein or nucleotide sequences. The basic local alignment search tool (blast) finds regions of local similarity between sequences. The program compares nucleotide or protein sequences to sequence. The program compares nucleotide or protein. Compares one or more protein query sequences to a. Blast Protein Comparison.
From www.pnas.org
Blast resistance of CCNBLRR protein Pb1 is mediated by WRKY45 through Blast Protein Comparison The program compares nucleotide or protein. The program compares nucleotide or protein sequences to sequence. Compares one or more protein query sequences to a subject protein sequence or a database of protein sequences. See also, the blast tutorial series:. This tutorial will explain how to compare and identify variation between two or more protein sequences. There are versions of blast.. Blast Protein Comparison.
From www.researchgate.net
Improved protein annotation via BLAST using MPAv2 in comparison to Blast Protein Comparison Blastp simply compares a protein query to a protein database. The program compares nucleotide or protein sequences to sequence. See also, the blast tutorial series:. A new approach to rapid sequence comparison, basic local alignment search tool (blast), directly approximates alignments that optimize a measure of local similarity, the maximal. The basic local alignment search tool (blast) finds regions of. Blast Protein Comparison.
From www.slideserve.com
PPT NCBI Molecular Biology Resources PowerPoint Presentation, free Blast Protein Comparison See also, the blast tutorial series:. There are versions of blast. This tutorial will explain how to compare and identify variation between two or more protein sequences. The program compares nucleotide or protein. Compares one or more protein query sequences to a subject protein sequence or a database of protein sequences. Blastp simply compares a protein query to a protein. Blast Protein Comparison.
From www.cancerdata.re.kr
Kcore Blast Protein Comparison The program compares nucleotide or protein. The basic local alignment search tool (blast) finds regions of local similarity between protein or nucleotide sequences. The basic local alignment search tool (blast) is the most widely used sequence similarity tool. The basic local alignment search tool (blast) finds regions of local similarity between sequences. The program compares nucleotide or protein sequences to. Blast Protein Comparison.
From www.foodandhealth.com
Protein Comparison PDF Printable Handout — Food and Health Communications Blast Protein Comparison The basic local alignment search tool (blast) finds regions of local similarity between protein or nucleotide sequences. The program compares nucleotide or protein. There are versions of blast. See also, the blast tutorial series:. A new approach to rapid sequence comparison, basic local alignment search tool (blast), directly approximates alignments that optimize a measure of local similarity, the maximal. The. Blast Protein Comparison.
From www.researchgate.net
A BLAST matrix of an all against all protein comparison of 12 Blast Protein Comparison Blastp simply compares a protein query to a protein database. There are versions of blast. The basic local alignment search tool (blast) finds regions of local similarity between sequences. This tutorial will explain how to compare and identify variation between two or more protein sequences. See also, the blast tutorial series:. This is useful when trying to. The program compares. Blast Protein Comparison.
From www.collegenutritionist.com
Ultimate Protein Comparison Blast Protein Comparison A new approach to rapid sequence comparison, basic local alignment search tool (blast), directly approximates alignments that optimize a measure of local similarity, the maximal. The program compares nucleotide or protein. The basic local alignment search tool (blast) finds regions of local similarity between sequences. Compares one or more protein query sequences to a subject protein sequence or a database. Blast Protein Comparison.
From www.foodandhealth.com
Protein Comparison PDF Printable Handout — Food and Health Communications Blast Protein Comparison This is useful when trying to. The program compares nucleotide or protein. See also, the blast tutorial series:. The basic local alignment search tool (blast) finds regions of local similarity between sequences. A new approach to rapid sequence comparison, basic local alignment search tool (blast), directly approximates alignments that optimize a measure of local similarity, the maximal. The basic local. Blast Protein Comparison.
From sequenceserver.com
Identifying gene sequences in a new genome assembly using BLAST Blast Protein Comparison The program compares nucleotide or protein sequences to sequence. A new approach to rapid sequence comparison, basic local alignment search tool (blast), directly approximates alignments that optimize a measure of local similarity, the maximal. Compares one or more protein query sequences to a subject protein sequence or a database of protein sequences. The basic local alignment search tool (blast) is. Blast Protein Comparison.
From www.semanticscholar.org
Figure 1 from BLAST 2 Sequences, a new tool for comparing protein and Blast Protein Comparison The program compares nucleotide or protein sequences to sequence. This tutorial will explain how to compare and identify variation between two or more protein sequences. The basic local alignment search tool (blast) finds regions of local similarity between sequences. The basic local alignment search tool (blast) is the most widely used sequence similarity tool. There are versions of blast. A. Blast Protein Comparison.
From www.researchgate.net
Protein blast similarity and secondary structure details of selected Blast Protein Comparison Blastp simply compares a protein query to a protein database. A new approach to rapid sequence comparison, basic local alignment search tool (blast), directly approximates alignments that optimize a measure of local similarity, the maximal. The basic local alignment search tool (blast) is the most widely used sequence similarity tool. Compares one or more protein query sequences to a subject. Blast Protein Comparison.
From www.researchgate.net
Results of protein sequence comparison, by the exact match and BLAST Blast Protein Comparison The basic local alignment search tool (blast) is the most widely used sequence similarity tool. A new approach to rapid sequence comparison, basic local alignment search tool (blast), directly approximates alignments that optimize a measure of local similarity, the maximal. The program compares nucleotide or protein sequences to sequence. There are versions of blast. The program compares nucleotide or protein.. Blast Protein Comparison.
From www.researchgate.net
A comparison of HMMER and BLAST programs for protein sequence analysis Blast Protein Comparison Blastp simply compares a protein query to a protein database. A new approach to rapid sequence comparison, basic local alignment search tool (blast), directly approximates alignments that optimize a measure of local similarity, the maximal. There are versions of blast. The basic local alignment search tool (blast) finds regions of local similarity between protein or nucleotide sequences. This tutorial will. Blast Protein Comparison.
From fity.club
Blastn Blast Protein Comparison The program compares nucleotide or protein sequences to sequence. Blastp simply compares a protein query to a protein database. The basic local alignment search tool (blast) finds regions of local similarity between sequences. The program compares nucleotide or protein. The basic local alignment search tool (blast) is the most widely used sequence similarity tool. This tutorial will explain how to. Blast Protein Comparison.
From www.slideserve.com
PPT Introduction to BLAST PowerPoint Presentation, free download ID Blast Protein Comparison A new approach to rapid sequence comparison, basic local alignment search tool (blast), directly approximates alignments that optimize a measure of local similarity, the maximal. The basic local alignment search tool (blast) finds regions of local similarity between sequences. This tutorial will explain how to compare and identify variation between two or more protein sequences. The basic local alignment search. Blast Protein Comparison.
From www.youtube.com
Which BLAST Algorithm to use for ProteinProtein BLAST YouTube Blast Protein Comparison See also, the blast tutorial series:. The basic local alignment search tool (blast) is the most widely used sequence similarity tool. This tutorial will explain how to compare and identify variation between two or more protein sequences. There are versions of blast. A new approach to rapid sequence comparison, basic local alignment search tool (blast), directly approximates alignments that optimize. Blast Protein Comparison.
From www.researchgate.net
a Hits returned from a protein blast search using eeAChE as a query. b Blast Protein Comparison Compares one or more protein query sequences to a subject protein sequence or a database of protein sequences. The basic local alignment search tool (blast) is the most widely used sequence similarity tool. Blastp simply compares a protein query to a protein database. The basic local alignment search tool (blast) finds regions of local similarity between sequences. The basic local. Blast Protein Comparison.
From www.youtube.com
Protein to Translated Nucleotide BLAST tBLASTn YouTube Blast Protein Comparison A new approach to rapid sequence comparison, basic local alignment search tool (blast), directly approximates alignments that optimize a measure of local similarity, the maximal. The program compares nucleotide or protein. The program compares nucleotide or protein sequences to sequence. The basic local alignment search tool (blast) finds regions of local similarity between protein or nucleotide sequences. This is useful. Blast Protein Comparison.
From gioxdsisl.blob.core.windows.net
Proteins For Blast at William Frady blog Blast Protein Comparison Compares one or more protein query sequences to a subject protein sequence or a database of protein sequences. The basic local alignment search tool (blast) is the most widely used sequence similarity tool. This is useful when trying to. A new approach to rapid sequence comparison, basic local alignment search tool (blast), directly approximates alignments that optimize a measure of. Blast Protein Comparison.
From www.youtube.com
Which Database to Choose for Protein BLAST run YouTube Blast Protein Comparison This tutorial will explain how to compare and identify variation between two or more protein sequences. The basic local alignment search tool (blast) finds regions of local similarity between protein or nucleotide sequences. The program compares nucleotide or protein sequences to sequence. The program compares nucleotide or protein. A new approach to rapid sequence comparison, basic local alignment search tool. Blast Protein Comparison.
From www.slideserve.com
PPT BLAST PowerPoint Presentation ID3793287 Blast Protein Comparison The basic local alignment search tool (blast) is the most widely used sequence similarity tool. There are versions of blast. A new approach to rapid sequence comparison, basic local alignment search tool (blast), directly approximates alignments that optimize a measure of local similarity, the maximal. The basic local alignment search tool (blast) finds regions of local similarity between protein or. Blast Protein Comparison.