Blast Protein Query . The basic local alignment search tool (blast) finds regions of local similarity between protein or nucleotide sequences. The basic local alignment search tool (blast) is the most widely used sequence similarity tool. The basic local alignment search tool (blast) finds regions of local similarity between sequences. A blast search enables a researcher to compare a subject protein (called a query) with a database of sequences, and identify database sequences. Tblastn compares a protein query sequence to all six possible reading frames of a database and is often used to identify proteins in new,. There are versions of blast. Quickblastp is an accelerated version of blastp that is very fast and works best if the target percent identity is 50% or more. The basic local alignment search tool (blast) finds regions of local similarity between protein or nucleotide. The program compares nucleotide or protein. Compares one or more protein query sequences to a subject protein sequence or a.
from chanzuckerberg.zendesk.com
A blast search enables a researcher to compare a subject protein (called a query) with a database of sequences, and identify database sequences. The program compares nucleotide or protein. The basic local alignment search tool (blast) finds regions of local similarity between protein or nucleotide sequences. The basic local alignment search tool (blast) finds regions of local similarity between protein or nucleotide. Compares one or more protein query sequences to a subject protein sequence or a. Quickblastp is an accelerated version of blastp that is very fast and works best if the target percent identity is 50% or more. The basic local alignment search tool (blast) is the most widely used sequence similarity tool. There are versions of blast. The basic local alignment search tool (blast) finds regions of local similarity between sequences. Tblastn compares a protein query sequence to all six possible reading frames of a database and is often used to identify proteins in new,.
A Guide to BLAST CZ ID Help Center
Blast Protein Query Tblastn compares a protein query sequence to all six possible reading frames of a database and is often used to identify proteins in new,. The basic local alignment search tool (blast) finds regions of local similarity between protein or nucleotide. The program compares nucleotide or protein. Compares one or more protein query sequences to a subject protein sequence or a. Tblastn compares a protein query sequence to all six possible reading frames of a database and is often used to identify proteins in new,. The basic local alignment search tool (blast) finds regions of local similarity between protein or nucleotide sequences. A blast search enables a researcher to compare a subject protein (called a query) with a database of sequences, and identify database sequences. There are versions of blast. Quickblastp is an accelerated version of blastp that is very fast and works best if the target percent identity is 50% or more. The basic local alignment search tool (blast) is the most widely used sequence similarity tool. The basic local alignment search tool (blast) finds regions of local similarity between sequences.
From www.slideserve.com
PPT BLAST PowerPoint Presentation, free download ID3794041 Blast Protein Query The basic local alignment search tool (blast) is the most widely used sequence similarity tool. A blast search enables a researcher to compare a subject protein (called a query) with a database of sequences, and identify database sequences. There are versions of blast. Quickblastp is an accelerated version of blastp that is very fast and works best if the target. Blast Protein Query.
From www.ncbi.nlm.nih.gov
Figure 1, The new BLAST output for a multiple sequence blastx Blast Protein Query Quickblastp is an accelerated version of blastp that is very fast and works best if the target percent identity is 50% or more. There are versions of blast. The basic local alignment search tool (blast) is the most widely used sequence similarity tool. Tblastn compares a protein query sequence to all six possible reading frames of a database and is. Blast Protein Query.
From www.youtube.com
How to Use BLAST for Finding and Aligning DNA or Protein Sequences Blast Protein Query Compares one or more protein query sequences to a subject protein sequence or a. The basic local alignment search tool (blast) finds regions of local similarity between protein or nucleotide sequences. Tblastn compares a protein query sequence to all six possible reading frames of a database and is often used to identify proteins in new,. The basic local alignment search. Blast Protein Query.
From www.researchgate.net
The BLAST algorithm. (a) Given a query sequence of length L, BLAST Blast Protein Query The basic local alignment search tool (blast) is the most widely used sequence similarity tool. The basic local alignment search tool (blast) finds regions of local similarity between protein or nucleotide. There are versions of blast. Compares one or more protein query sequences to a subject protein sequence or a. Quickblastp is an accelerated version of blastp that is very. Blast Protein Query.
From tcdb.org
TCBLAST Tutorial Blast Protein Query Compares one or more protein query sequences to a subject protein sequence or a. The basic local alignment search tool (blast) is the most widely used sequence similarity tool. Tblastn compares a protein query sequence to all six possible reading frames of a database and is often used to identify proteins in new,. The basic local alignment search tool (blast). Blast Protein Query.
From gioxdsisl.blob.core.windows.net
Proteins For Blast at William Frady blog Blast Protein Query There are versions of blast. The basic local alignment search tool (blast) is the most widely used sequence similarity tool. The program compares nucleotide or protein. A blast search enables a researcher to compare a subject protein (called a query) with a database of sequences, and identify database sequences. Tblastn compares a protein query sequence to all six possible reading. Blast Protein Query.
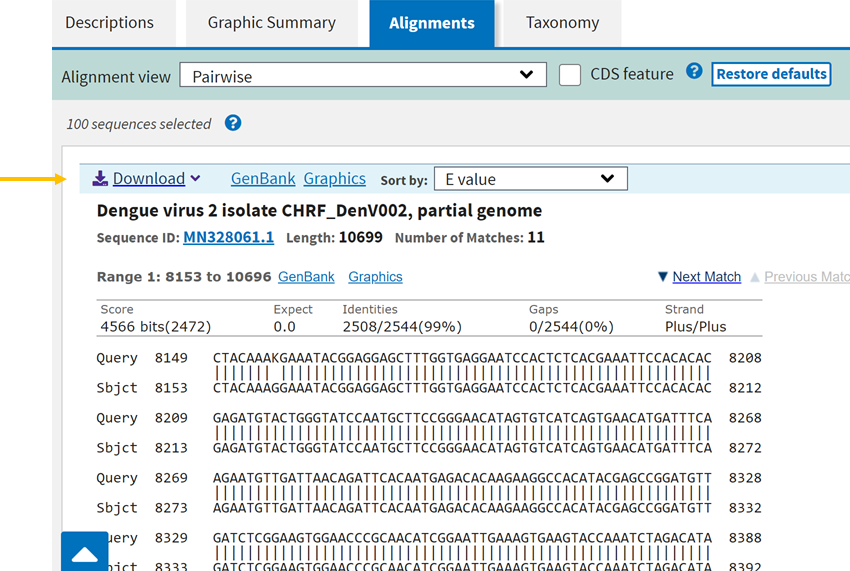
From support.nlm.nih.gov
How do I use Nucleotide BLAST (blastn) and CDS feature display to Blast Protein Query The program compares nucleotide or protein. The basic local alignment search tool (blast) is the most widely used sequence similarity tool. The basic local alignment search tool (blast) finds regions of local similarity between protein or nucleotide. The basic local alignment search tool (blast) finds regions of local similarity between protein or nucleotide sequences. A blast search enables a researcher. Blast Protein Query.
From bch709.plantgenomicslab.org
HPC_RNA_Seq BCH709 Introduction to Bioinformatics Blast Protein Query The basic local alignment search tool (blast) finds regions of local similarity between protein or nucleotide sequences. Compares one or more protein query sequences to a subject protein sequence or a. The basic local alignment search tool (blast) is the most widely used sequence similarity tool. Quickblastp is an accelerated version of blastp that is very fast and works best. Blast Protein Query.
From usuhs.libguides.com
BLAST Results Introduction to NCBI Bioinformatics Resources Blast Protein Query Tblastn compares a protein query sequence to all six possible reading frames of a database and is often used to identify proteins in new,. The basic local alignment search tool (blast) finds regions of local similarity between sequences. Compares one or more protein query sequences to a subject protein sequence or a. The program compares nucleotide or protein. There are. Blast Protein Query.
From www.slideserve.com
PPT BLAST PowerPoint Presentation ID3793287 Blast Protein Query A blast search enables a researcher to compare a subject protein (called a query) with a database of sequences, and identify database sequences. The program compares nucleotide or protein. The basic local alignment search tool (blast) finds regions of local similarity between sequences. The basic local alignment search tool (blast) finds regions of local similarity between protein or nucleotide sequences.. Blast Protein Query.
From www.researchgate.net
NCBI Conserved Domains annotated on a protein query. (a) RPSBLAST is Blast Protein Query A blast search enables a researcher to compare a subject protein (called a query) with a database of sequences, and identify database sequences. Quickblastp is an accelerated version of blastp that is very fast and works best if the target percent identity is 50% or more. The basic local alignment search tool (blast) is the most widely used sequence similarity. Blast Protein Query.
From www.slideserve.com
PPT Database Searches PowerPoint Presentation, free download ID1439229 Blast Protein Query The basic local alignment search tool (blast) finds regions of local similarity between protein or nucleotide sequences. The basic local alignment search tool (blast) finds regions of local similarity between protein or nucleotide. The basic local alignment search tool (blast) finds regions of local similarity between sequences. Quickblastp is an accelerated version of blastp that is very fast and works. Blast Protein Query.
From www.researchgate.net
List of NCBI database protein accession numbers used in the BLASTp Blast Protein Query Tblastn compares a protein query sequence to all six possible reading frames of a database and is often used to identify proteins in new,. The basic local alignment search tool (blast) finds regions of local similarity between sequences. The basic local alignment search tool (blast) is the most widely used sequence similarity tool. A blast search enables a researcher to. Blast Protein Query.
From studylib.net
Where to find protein sequences Blast Protein Query The program compares nucleotide or protein. Quickblastp is an accelerated version of blastp that is very fast and works best if the target percent identity is 50% or more. There are versions of blast. The basic local alignment search tool (blast) finds regions of local similarity between protein or nucleotide sequences. Tblastn compares a protein query sequence to all six. Blast Protein Query.
From www.researchgate.net
The blast query form allows to search the contigs as a database using Blast Protein Query The basic local alignment search tool (blast) finds regions of local similarity between protein or nucleotide. Tblastn compares a protein query sequence to all six possible reading frames of a database and is often used to identify proteins in new,. The basic local alignment search tool (blast) finds regions of local similarity between protein or nucleotide sequences. A blast search. Blast Protein Query.
From www.researchgate.net
A protein sequence query and results. Download Scientific Diagram Blast Protein Query The basic local alignment search tool (blast) finds regions of local similarity between protein or nucleotide sequences. Quickblastp is an accelerated version of blastp that is very fast and works best if the target percent identity is 50% or more. Compares one or more protein query sequences to a subject protein sequence or a. The basic local alignment search tool. Blast Protein Query.
From usuhs.libguides.com
BLAST Results Introduction to NCBI Bioinformatics Resources Blast Protein Query The basic local alignment search tool (blast) is the most widely used sequence similarity tool. The program compares nucleotide or protein. A blast search enables a researcher to compare a subject protein (called a query) with a database of sequences, and identify database sequences. The basic local alignment search tool (blast) finds regions of local similarity between sequences. The basic. Blast Protein Query.
From www.researchgate.net
NCBI BLAST search of the assembled sequence of Arthroleptis Blast Protein Query The basic local alignment search tool (blast) finds regions of local similarity between sequences. The basic local alignment search tool (blast) finds regions of local similarity between protein or nucleotide. A blast search enables a researcher to compare a subject protein (called a query) with a database of sequences, and identify database sequences. The basic local alignment search tool (blast). Blast Protein Query.
From sgbc.github.io
Blast Online Bioinformatics course Blast Protein Query There are versions of blast. The basic local alignment search tool (blast) is the most widely used sequence similarity tool. The program compares nucleotide or protein. The basic local alignment search tool (blast) finds regions of local similarity between protein or nucleotide. A blast search enables a researcher to compare a subject protein (called a query) with a database of. Blast Protein Query.
From slideplayer.com
SEQUENCE ANALYSIS By Jyotika Bhati. ppt video online download Blast Protein Query The basic local alignment search tool (blast) is the most widely used sequence similarity tool. The basic local alignment search tool (blast) finds regions of local similarity between protein or nucleotide sequences. A blast search enables a researcher to compare a subject protein (called a query) with a database of sequences, and identify database sequences. Quickblastp is an accelerated version. Blast Protein Query.
From www.researchgate.net
NCBI Conserved Domains annotated on a protein query. (a) RPSBLAST is Blast Protein Query A blast search enables a researcher to compare a subject protein (called a query) with a database of sequences, and identify database sequences. The basic local alignment search tool (blast) is the most widely used sequence similarity tool. The basic local alignment search tool (blast) finds regions of local similarity between protein or nucleotide sequences. The basic local alignment search. Blast Protein Query.
From www.slideserve.com
PPT BLAST Basic Local Alignment Search Tool PowerPoint Presentation Blast Protein Query The program compares nucleotide or protein. Tblastn compares a protein query sequence to all six possible reading frames of a database and is often used to identify proteins in new,. The basic local alignment search tool (blast) is the most widely used sequence similarity tool. Compares one or more protein query sequences to a subject protein sequence or a. The. Blast Protein Query.
From usuhs.libguides.com
BLAST Results Introduction to NCBI Bioinformatics Resources Blast Protein Query A blast search enables a researcher to compare a subject protein (called a query) with a database of sequences, and identify database sequences. The basic local alignment search tool (blast) finds regions of local similarity between protein or nucleotide sequences. The basic local alignment search tool (blast) finds regions of local similarity between sequences. Quickblastp is an accelerated version of. Blast Protein Query.
From chanzuckerberg.zendesk.com
A Guide to BLAST CZ ID Help Center Blast Protein Query Quickblastp is an accelerated version of blastp that is very fast and works best if the target percent identity is 50% or more. The basic local alignment search tool (blast) finds regions of local similarity between protein or nucleotide. Tblastn compares a protein query sequence to all six possible reading frames of a database and is often used to identify. Blast Protein Query.
From www.slideserve.com
PPT CS 6990 Bioinformatics BLAST PowerPoint Presentation, free Blast Protein Query The basic local alignment search tool (blast) finds regions of local similarity between protein or nucleotide. The basic local alignment search tool (blast) finds regions of local similarity between protein or nucleotide sequences. There are versions of blast. The program compares nucleotide or protein. Tblastn compares a protein query sequence to all six possible reading frames of a database and. Blast Protein Query.
From tcdb.org
TCBLAST Tutorial Blast Protein Query The program compares nucleotide or protein. The basic local alignment search tool (blast) finds regions of local similarity between sequences. The basic local alignment search tool (blast) finds regions of local similarity between protein or nucleotide sequences. A blast search enables a researcher to compare a subject protein (called a query) with a database of sequences, and identify database sequences.. Blast Protein Query.
From slideplayer.com
Predicting protein structure and function ppt download Blast Protein Query Tblastn compares a protein query sequence to all six possible reading frames of a database and is often used to identify proteins in new,. The basic local alignment search tool (blast) is the most widely used sequence similarity tool. Quickblastp is an accelerated version of blastp that is very fast and works best if the target percent identity is 50%. Blast Protein Query.
From slidetodoc.com
BLAST Sequence alignment Evalue Extreme value distribution Database Blast Protein Query There are versions of blast. The basic local alignment search tool (blast) finds regions of local similarity between sequences. The basic local alignment search tool (blast) finds regions of local similarity between protein or nucleotide. The basic local alignment search tool (blast) is the most widely used sequence similarity tool. Quickblastp is an accelerated version of blastp that is very. Blast Protein Query.
From www.slideserve.com
PPT BLAST PowerPoint Presentation, free download ID3384708 Blast Protein Query The program compares nucleotide or protein. Tblastn compares a protein query sequence to all six possible reading frames of a database and is often used to identify proteins in new,. There are versions of blast. Quickblastp is an accelerated version of blastp that is very fast and works best if the target percent identity is 50% or more. The basic. Blast Protein Query.
From www.slideserve.com
PPT BLAST PowerPoint Presentation, free download ID3793287 Blast Protein Query The basic local alignment search tool (blast) finds regions of local similarity between sequences. The basic local alignment search tool (blast) is the most widely used sequence similarity tool. The program compares nucleotide or protein. The basic local alignment search tool (blast) finds regions of local similarity between protein or nucleotide. Tblastn compares a protein query sequence to all six. Blast Protein Query.
From abacus.bates.edu
BLASTP Results Part 1 Blast Protein Query There are versions of blast. The basic local alignment search tool (blast) finds regions of local similarity between sequences. The basic local alignment search tool (blast) finds regions of local similarity between protein or nucleotide sequences. Quickblastp is an accelerated version of blastp that is very fast and works best if the target percent identity is 50% or more. Compares. Blast Protein Query.
From www.slideserve.com
PPT Basic Introduction of BLAST PowerPoint Presentation, free Blast Protein Query Compares one or more protein query sequences to a subject protein sequence or a. Quickblastp is an accelerated version of blastp that is very fast and works best if the target percent identity is 50% or more. The basic local alignment search tool (blast) is the most widely used sequence similarity tool. The program compares nucleotide or protein. A blast. Blast Protein Query.
From www.researchgate.net
BLAST outputs of total score, query coverage, evalue, percentage Blast Protein Query The basic local alignment search tool (blast) is the most widely used sequence similarity tool. The basic local alignment search tool (blast) finds regions of local similarity between protein or nucleotide. Compares one or more protein query sequences to a subject protein sequence or a. There are versions of blast. The basic local alignment search tool (blast) finds regions of. Blast Protein Query.
From www.researchgate.net
List of NCBI database protein accession numbers used in the BLASTp Blast Protein Query There are versions of blast. The program compares nucleotide or protein. The basic local alignment search tool (blast) is the most widely used sequence similarity tool. The basic local alignment search tool (blast) finds regions of local similarity between protein or nucleotide sequences. Tblastn compares a protein query sequence to all six possible reading frames of a database and is. Blast Protein Query.
From www.slideserve.com
PPT FASTA and BLAST PowerPoint Presentation ID948138 Blast Protein Query Compares one or more protein query sequences to a subject protein sequence or a. The basic local alignment search tool (blast) is the most widely used sequence similarity tool. Tblastn compares a protein query sequence to all six possible reading frames of a database and is often used to identify proteins in new,. The program compares nucleotide or protein. A. Blast Protein Query.