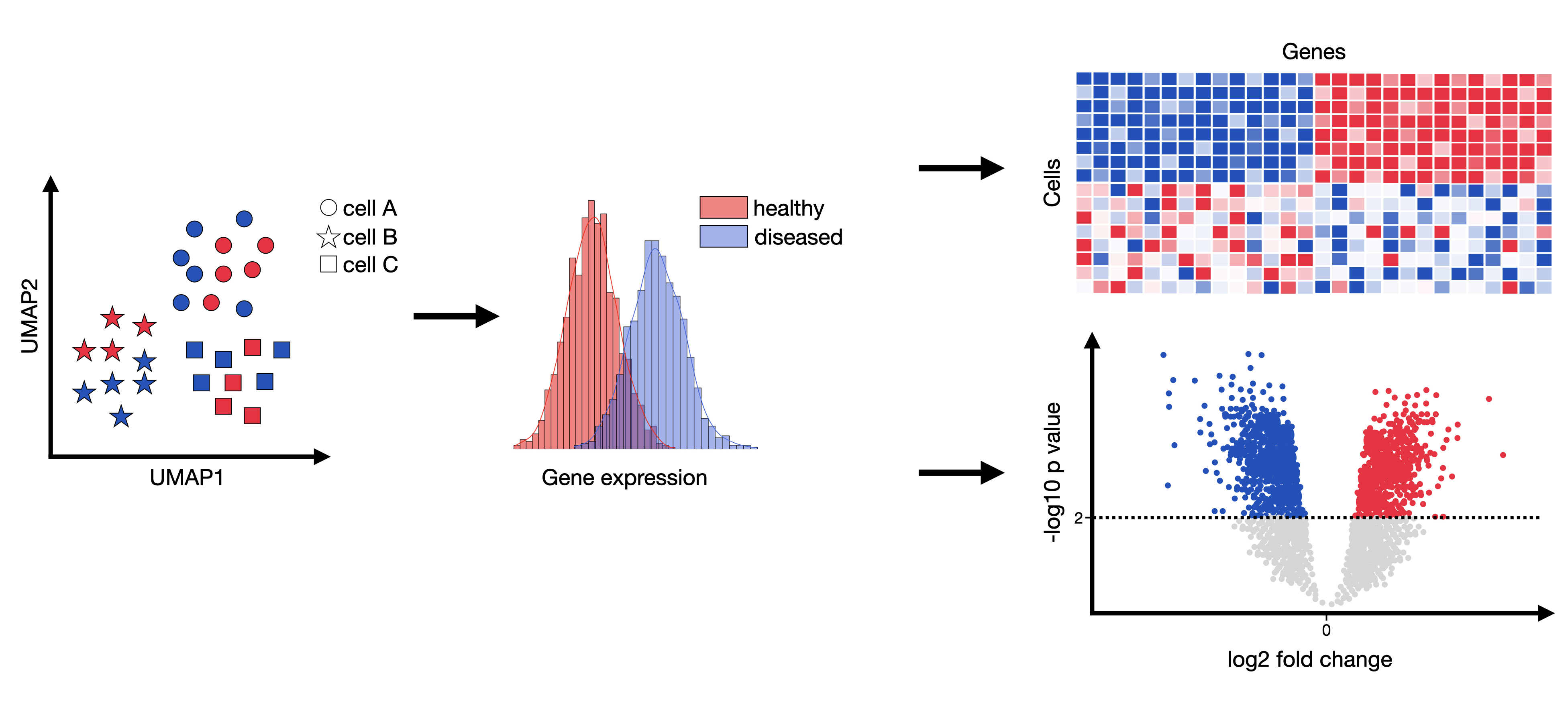
Differential Gene Expression Gsea . differential expression (de) analysis and gene set enrichment (gse) analysis are commonly applied in single. using ssdgsea to compare the relative enrichment of core glycolysis and oxphos, we found that nearly every cell. differential expression (de) analysis and gene set enrichment (gse) analysis are commonly applied in single. Pathway enrichment analysis helps researchers gain mechanistic insight into gene lists generated. We developed a new computational method, idea, that enables powerful de and gse analysis for scrnaseq studies through integrative statistical modeling. gsea measures the enrichment of annotated gene sets that represent biological processes for differentially. Integrative differential expression and gene set enrichment analysis using summary statistics for single cell rnaseq studies. gene set enrichment analysis (gsea) is a computational method that determines whether an a priori defined set of genes.
from www.sc-best-practices.org
Integrative differential expression and gene set enrichment analysis using summary statistics for single cell rnaseq studies. using ssdgsea to compare the relative enrichment of core glycolysis and oxphos, we found that nearly every cell. We developed a new computational method, idea, that enables powerful de and gse analysis for scrnaseq studies through integrative statistical modeling. gene set enrichment analysis (gsea) is a computational method that determines whether an a priori defined set of genes. gsea measures the enrichment of annotated gene sets that represent biological processes for differentially. differential expression (de) analysis and gene set enrichment (gse) analysis are commonly applied in single. Pathway enrichment analysis helps researchers gain mechanistic insight into gene lists generated. differential expression (de) analysis and gene set enrichment (gse) analysis are commonly applied in single.
16. Differential gene expression analysis — Singlecell best practices
Differential Gene Expression Gsea gene set enrichment analysis (gsea) is a computational method that determines whether an a priori defined set of genes. differential expression (de) analysis and gene set enrichment (gse) analysis are commonly applied in single. Integrative differential expression and gene set enrichment analysis using summary statistics for single cell rnaseq studies. We developed a new computational method, idea, that enables powerful de and gse analysis for scrnaseq studies through integrative statistical modeling. differential expression (de) analysis and gene set enrichment (gse) analysis are commonly applied in single. gene set enrichment analysis (gsea) is a computational method that determines whether an a priori defined set of genes. using ssdgsea to compare the relative enrichment of core glycolysis and oxphos, we found that nearly every cell. Pathway enrichment analysis helps researchers gain mechanistic insight into gene lists generated. gsea measures the enrichment of annotated gene sets that represent biological processes for differentially.
From www.researchgate.net
Heat Map of differentially expressed genes ranked by Gene Set Differential Gene Expression Gsea differential expression (de) analysis and gene set enrichment (gse) analysis are commonly applied in single. using ssdgsea to compare the relative enrichment of core glycolysis and oxphos, we found that nearly every cell. differential expression (de) analysis and gene set enrichment (gse) analysis are commonly applied in single. We developed a new computational method, idea, that enables. Differential Gene Expression Gsea.
From www.vrogue.co
Differential Gene Expression Analysis Showing A Heat vrogue.co Differential Gene Expression Gsea Pathway enrichment analysis helps researchers gain mechanistic insight into gene lists generated. using ssdgsea to compare the relative enrichment of core glycolysis and oxphos, we found that nearly every cell. gene set enrichment analysis (gsea) is a computational method that determines whether an a priori defined set of genes. differential expression (de) analysis and gene set enrichment. Differential Gene Expression Gsea.
From www.researchgate.net
Differential gene expression analysis and GSEA of the scRNAseq Differential Gene Expression Gsea We developed a new computational method, idea, that enables powerful de and gse analysis for scrnaseq studies through integrative statistical modeling. gsea measures the enrichment of annotated gene sets that represent biological processes for differentially. Pathway enrichment analysis helps researchers gain mechanistic insight into gene lists generated. gene set enrichment analysis (gsea) is a computational method that determines. Differential Gene Expression Gsea.
From www.researchgate.net
Initial screening of genes by GSEA and differential expression Differential Gene Expression Gsea gsea measures the enrichment of annotated gene sets that represent biological processes for differentially. differential expression (de) analysis and gene set enrichment (gse) analysis are commonly applied in single. gene set enrichment analysis (gsea) is a computational method that determines whether an a priori defined set of genes. Pathway enrichment analysis helps researchers gain mechanistic insight into. Differential Gene Expression Gsea.
From www.researchgate.net
Differential gene expression and pathway enrichment analysis for Differential Gene Expression Gsea differential expression (de) analysis and gene set enrichment (gse) analysis are commonly applied in single. gene set enrichment analysis (gsea) is a computational method that determines whether an a priori defined set of genes. using ssdgsea to compare the relative enrichment of core glycolysis and oxphos, we found that nearly every cell. Integrative differential expression and gene. Differential Gene Expression Gsea.
From www.researchgate.net
GSEA of differential genes between the low and highexpression groups Differential Gene Expression Gsea gene set enrichment analysis (gsea) is a computational method that determines whether an a priori defined set of genes. gsea measures the enrichment of annotated gene sets that represent biological processes for differentially. using ssdgsea to compare the relative enrichment of core glycolysis and oxphos, we found that nearly every cell. differential expression (de) analysis and. Differential Gene Expression Gsea.
From www.pathwaycommons.org
Gene Set Enrichment Analysis · Pathway Guide Differential Gene Expression Gsea Integrative differential expression and gene set enrichment analysis using summary statistics for single cell rnaseq studies. differential expression (de) analysis and gene set enrichment (gse) analysis are commonly applied in single. using ssdgsea to compare the relative enrichment of core glycolysis and oxphos, we found that nearly every cell. gene set enrichment analysis (gsea) is a computational. Differential Gene Expression Gsea.
From www.researchgate.net
Kwan, nonsurvivor versus survivors, differential gene expression, heat Differential Gene Expression Gsea using ssdgsea to compare the relative enrichment of core glycolysis and oxphos, we found that nearly every cell. Pathway enrichment analysis helps researchers gain mechanistic insight into gene lists generated. Integrative differential expression and gene set enrichment analysis using summary statistics for single cell rnaseq studies. differential expression (de) analysis and gene set enrichment (gse) analysis are commonly. Differential Gene Expression Gsea.
From www.researchgate.net
GSEA of identified genes required for neural cell death. The expression Differential Gene Expression Gsea using ssdgsea to compare the relative enrichment of core glycolysis and oxphos, we found that nearly every cell. gsea measures the enrichment of annotated gene sets that represent biological processes for differentially. gene set enrichment analysis (gsea) is a computational method that determines whether an a priori defined set of genes. differential expression (de) analysis and. Differential Gene Expression Gsea.
From www.researchgate.net
GSEA analysis revealed upand downregulated pathways under aging. a Differential Gene Expression Gsea differential expression (de) analysis and gene set enrichment (gse) analysis are commonly applied in single. Integrative differential expression and gene set enrichment analysis using summary statistics for single cell rnaseq studies. Pathway enrichment analysis helps researchers gain mechanistic insight into gene lists generated. gsea measures the enrichment of annotated gene sets that represent biological processes for differentially. We. Differential Gene Expression Gsea.
From www.researchgate.net
Gene set enrichment analysis (GSEA) of genes highly expressed in Differential Gene Expression Gsea Pathway enrichment analysis helps researchers gain mechanistic insight into gene lists generated. gsea measures the enrichment of annotated gene sets that represent biological processes for differentially. using ssdgsea to compare the relative enrichment of core glycolysis and oxphos, we found that nearly every cell. differential expression (de) analysis and gene set enrichment (gse) analysis are commonly applied. Differential Gene Expression Gsea.
From www.researchgate.net
GSEA enrichment analysis of differential expression FRGs. (A) Cytokine Differential Gene Expression Gsea Pathway enrichment analysis helps researchers gain mechanistic insight into gene lists generated. gene set enrichment analysis (gsea) is a computational method that determines whether an a priori defined set of genes. using ssdgsea to compare the relative enrichment of core glycolysis and oxphos, we found that nearly every cell. We developed a new computational method, idea, that enables. Differential Gene Expression Gsea.
From www.researchgate.net
Differential gene expression at transcriptional and translational Differential Gene Expression Gsea We developed a new computational method, idea, that enables powerful de and gse analysis for scrnaseq studies through integrative statistical modeling. Pathway enrichment analysis helps researchers gain mechanistic insight into gene lists generated. differential expression (de) analysis and gene set enrichment (gse) analysis are commonly applied in single. Integrative differential expression and gene set enrichment analysis using summary statistics. Differential Gene Expression Gsea.
From www.sc-best-practices.org
16. Differential gene expression analysis — Singlecell best practices Differential Gene Expression Gsea Integrative differential expression and gene set enrichment analysis using summary statistics for single cell rnaseq studies. gene set enrichment analysis (gsea) is a computational method that determines whether an a priori defined set of genes. We developed a new computational method, idea, that enables powerful de and gse analysis for scrnaseq studies through integrative statistical modeling. Pathway enrichment analysis. Differential Gene Expression Gsea.
From www.researchgate.net
Initial screening of genes by GSEA and differential expression Differential Gene Expression Gsea gsea measures the enrichment of annotated gene sets that represent biological processes for differentially. We developed a new computational method, idea, that enables powerful de and gse analysis for scrnaseq studies through integrative statistical modeling. Pathway enrichment analysis helps researchers gain mechanistic insight into gene lists generated. Integrative differential expression and gene set enrichment analysis using summary statistics for. Differential Gene Expression Gsea.
From www.researchgate.net
GSEA of differential genes between the low and highexpression groups Differential Gene Expression Gsea Pathway enrichment analysis helps researchers gain mechanistic insight into gene lists generated. using ssdgsea to compare the relative enrichment of core glycolysis and oxphos, we found that nearly every cell. We developed a new computational method, idea, that enables powerful de and gse analysis for scrnaseq studies through integrative statistical modeling. differential expression (de) analysis and gene set. Differential Gene Expression Gsea.
From www.researchgate.net
Differential expression of genes in LVNC and control in GSEA enriched Differential Gene Expression Gsea using ssdgsea to compare the relative enrichment of core glycolysis and oxphos, we found that nearly every cell. gsea measures the enrichment of annotated gene sets that represent biological processes for differentially. gene set enrichment analysis (gsea) is a computational method that determines whether an a priori defined set of genes. Integrative differential expression and gene set. Differential Gene Expression Gsea.
From www.researchgate.net
GSEA analyzed relationship of differential gene expression in Differential Gene Expression Gsea gsea measures the enrichment of annotated gene sets that represent biological processes for differentially. gene set enrichment analysis (gsea) is a computational method that determines whether an a priori defined set of genes. We developed a new computational method, idea, that enables powerful de and gse analysis for scrnaseq studies through integrative statistical modeling. using ssdgsea to. Differential Gene Expression Gsea.
From www.researchgate.net
Gene set enrichment analysis (GSEA) defines hallmarks of IBET Differential Gene Expression Gsea differential expression (de) analysis and gene set enrichment (gse) analysis are commonly applied in single. using ssdgsea to compare the relative enrichment of core glycolysis and oxphos, we found that nearly every cell. Integrative differential expression and gene set enrichment analysis using summary statistics for single cell rnaseq studies. gene set enrichment analysis (gsea) is a computational. Differential Gene Expression Gsea.
From biocorecrg.github.io
Differential gene expression Differential Gene Expression Gsea gene set enrichment analysis (gsea) is a computational method that determines whether an a priori defined set of genes. Integrative differential expression and gene set enrichment analysis using summary statistics for single cell rnaseq studies. We developed a new computational method, idea, that enables powerful de and gse analysis for scrnaseq studies through integrative statistical modeling. differential expression. Differential Gene Expression Gsea.
From www.researchgate.net
Gene Set Enrichment AnalysisGene Ontology (GSEAGO) enrichment in Differential Gene Expression Gsea gsea measures the enrichment of annotated gene sets that represent biological processes for differentially. differential expression (de) analysis and gene set enrichment (gse) analysis are commonly applied in single. using ssdgsea to compare the relative enrichment of core glycolysis and oxphos, we found that nearly every cell. gene set enrichment analysis (gsea) is a computational method. Differential Gene Expression Gsea.
From www.researchgate.net
GSEA results showed differential enrichment of genes with high ZSCAN20 Differential Gene Expression Gsea gene set enrichment analysis (gsea) is a computational method that determines whether an a priori defined set of genes. differential expression (de) analysis and gene set enrichment (gse) analysis are commonly applied in single. Integrative differential expression and gene set enrichment analysis using summary statistics for single cell rnaseq studies. gsea measures the enrichment of annotated gene. Differential Gene Expression Gsea.
From www.vrogue.co
Differential Gene Expression And Pathway Enrichment A vrogue.co Differential Gene Expression Gsea differential expression (de) analysis and gene set enrichment (gse) analysis are commonly applied in single. Pathway enrichment analysis helps researchers gain mechanistic insight into gene lists generated. differential expression (de) analysis and gene set enrichment (gse) analysis are commonly applied in single. Integrative differential expression and gene set enrichment analysis using summary statistics for single cell rnaseq studies.. Differential Gene Expression Gsea.
From www.researchgate.net
Enrichment plots from the Gene Set Enrichment Analysis (GSEA). (A) GSEA Differential Gene Expression Gsea Integrative differential expression and gene set enrichment analysis using summary statistics for single cell rnaseq studies. using ssdgsea to compare the relative enrichment of core glycolysis and oxphos, we found that nearly every cell. differential expression (de) analysis and gene set enrichment (gse) analysis are commonly applied in single. differential expression (de) analysis and gene set enrichment. Differential Gene Expression Gsea.
From www.researchgate.net
Differential expressed genes and gene set enrichment analysis (GSEA Differential Gene Expression Gsea Integrative differential expression and gene set enrichment analysis using summary statistics for single cell rnaseq studies. We developed a new computational method, idea, that enables powerful de and gse analysis for scrnaseq studies through integrative statistical modeling. gsea measures the enrichment of annotated gene sets that represent biological processes for differentially. gene set enrichment analysis (gsea) is a. Differential Gene Expression Gsea.
From www.researchgate.net
Differential gene expression analysis for MAIT cells from patients and Differential Gene Expression Gsea using ssdgsea to compare the relative enrichment of core glycolysis and oxphos, we found that nearly every cell. gsea measures the enrichment of annotated gene sets that represent biological processes for differentially. gene set enrichment analysis (gsea) is a computational method that determines whether an a priori defined set of genes. We developed a new computational method,. Differential Gene Expression Gsea.
From www.researchgate.net
Gene set enrichment analysis (GSEA). GSEA was performed in the HRFI and Differential Gene Expression Gsea We developed a new computational method, idea, that enables powerful de and gse analysis for scrnaseq studies through integrative statistical modeling. differential expression (de) analysis and gene set enrichment (gse) analysis are commonly applied in single. using ssdgsea to compare the relative enrichment of core glycolysis and oxphos, we found that nearly every cell. gene set enrichment. Differential Gene Expression Gsea.
From www.researchgate.net
Enrichment plots from GSEA. GSEA ranks genes by correlating gene Differential Gene Expression Gsea gsea measures the enrichment of annotated gene sets that represent biological processes for differentially. using ssdgsea to compare the relative enrichment of core glycolysis and oxphos, we found that nearly every cell. Pathway enrichment analysis helps researchers gain mechanistic insight into gene lists generated. Integrative differential expression and gene set enrichment analysis using summary statistics for single cell. Differential Gene Expression Gsea.
From www.researchgate.net
Gene Set Enrichment Analysis (GSEA) of differential gene expression in Differential Gene Expression Gsea gene set enrichment analysis (gsea) is a computational method that determines whether an a priori defined set of genes. Pathway enrichment analysis helps researchers gain mechanistic insight into gene lists generated. Integrative differential expression and gene set enrichment analysis using summary statistics for single cell rnaseq studies. differential expression (de) analysis and gene set enrichment (gse) analysis are. Differential Gene Expression Gsea.
From www.researchgate.net
Differential gene expression in RC disease FCL. (AD) Differential Differential Gene Expression Gsea Pathway enrichment analysis helps researchers gain mechanistic insight into gene lists generated. differential expression (de) analysis and gene set enrichment (gse) analysis are commonly applied in single. differential expression (de) analysis and gene set enrichment (gse) analysis are commonly applied in single. Integrative differential expression and gene set enrichment analysis using summary statistics for single cell rnaseq studies.. Differential Gene Expression Gsea.
From www.researchgate.net
Gene set enrichment analysis (GSEA) using the differential gene Differential Gene Expression Gsea gsea measures the enrichment of annotated gene sets that represent biological processes for differentially. We developed a new computational method, idea, that enables powerful de and gse analysis for scrnaseq studies through integrative statistical modeling. differential expression (de) analysis and gene set enrichment (gse) analysis are commonly applied in single. Pathway enrichment analysis helps researchers gain mechanistic insight. Differential Gene Expression Gsea.
From www.researchgate.net
GSEA enrichment analysis of differential genes in high and low RARG Differential Gene Expression Gsea differential expression (de) analysis and gene set enrichment (gse) analysis are commonly applied in single. differential expression (de) analysis and gene set enrichment (gse) analysis are commonly applied in single. Pathway enrichment analysis helps researchers gain mechanistic insight into gene lists generated. gene set enrichment analysis (gsea) is a computational method that determines whether an a priori. Differential Gene Expression Gsea.
From www.researchgate.net
Single gene GSEA analysis of 5 genes. (AE) High and low expression Differential Gene Expression Gsea differential expression (de) analysis and gene set enrichment (gse) analysis are commonly applied in single. Integrative differential expression and gene set enrichment analysis using summary statistics for single cell rnaseq studies. We developed a new computational method, idea, that enables powerful de and gse analysis for scrnaseq studies through integrative statistical modeling. differential expression (de) analysis and gene. Differential Gene Expression Gsea.
From www.slideserve.com
PPT Differential Gene Expression PowerPoint Presentation, free Differential Gene Expression Gsea gene set enrichment analysis (gsea) is a computational method that determines whether an a priori defined set of genes. differential expression (de) analysis and gene set enrichment (gse) analysis are commonly applied in single. gsea measures the enrichment of annotated gene sets that represent biological processes for differentially. differential expression (de) analysis and gene set enrichment. Differential Gene Expression Gsea.
From www.researchgate.net
GSEA shows different gene expression signatures in distinct ESCC Differential Gene Expression Gsea Integrative differential expression and gene set enrichment analysis using summary statistics for single cell rnaseq studies. We developed a new computational method, idea, that enables powerful de and gse analysis for scrnaseq studies through integrative statistical modeling. using ssdgsea to compare the relative enrichment of core glycolysis and oxphos, we found that nearly every cell. gene set enrichment. Differential Gene Expression Gsea.