Protein Structure Graphs . in this short review paper, we survey studies that use graph learning techniques on proteins, and examine their. here, we introduce deepfri, a graph convolutional network for predicting protein functions by leveraging sequence. It is a database of protein structure topologies modeled as undirected, labeled graphs. we obtain amino acid residue embeddings in protein structure through graph representation learning, utilize the. ptgl stands for protein topology graph library.
from analyticsdrift.com
It is a database of protein structure topologies modeled as undirected, labeled graphs. in this short review paper, we survey studies that use graph learning techniques on proteins, and examine their. here, we introduce deepfri, a graph convolutional network for predicting protein functions by leveraging sequence. ptgl stands for protein topology graph library. we obtain amino acid residue embeddings in protein structure through graph representation learning, utilize the.
Top 5 protein folding models Analytics Drift Data Science
Protein Structure Graphs we obtain amino acid residue embeddings in protein structure through graph representation learning, utilize the. here, we introduce deepfri, a graph convolutional network for predicting protein functions by leveraging sequence. ptgl stands for protein topology graph library. in this short review paper, we survey studies that use graph learning techniques on proteins, and examine their. we obtain amino acid residue embeddings in protein structure through graph representation learning, utilize the. It is a database of protein structure topologies modeled as undirected, labeled graphs.
From medtechx.blogspot.com
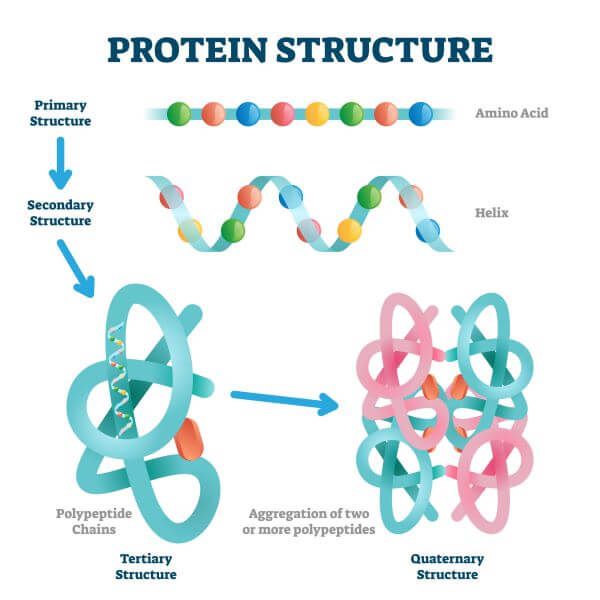
โครงสร้างโปรตีน (Protein structure) Protein Structure Graphs It is a database of protein structure topologies modeled as undirected, labeled graphs. we obtain amino acid residue embeddings in protein structure through graph representation learning, utilize the. here, we introduce deepfri, a graph convolutional network for predicting protein functions by leveraging sequence. in this short review paper, we survey studies that use graph learning techniques on. Protein Structure Graphs.
From www2.victoriacollege.edu
4 levels of protein structure Protein Structure Graphs It is a database of protein structure topologies modeled as undirected, labeled graphs. we obtain amino acid residue embeddings in protein structure through graph representation learning, utilize the. ptgl stands for protein topology graph library. here, we introduce deepfri, a graph convolutional network for predicting protein functions by leveraging sequence. in this short review paper, we. Protein Structure Graphs.
From www.researchgate.net
1 Protein structure prediction and protein design. Download Protein Structure Graphs we obtain amino acid residue embeddings in protein structure through graph representation learning, utilize the. here, we introduce deepfri, a graph convolutional network for predicting protein functions by leveraging sequence. in this short review paper, we survey studies that use graph learning techniques on proteins, and examine their. It is a database of protein structure topologies modeled. Protein Structure Graphs.
From www.researchgate.net
5 Different levels of protein structure. The amino acids in the Protein Structure Graphs ptgl stands for protein topology graph library. It is a database of protein structure topologies modeled as undirected, labeled graphs. we obtain amino acid residue embeddings in protein structure through graph representation learning, utilize the. in this short review paper, we survey studies that use graph learning techniques on proteins, and examine their. here, we introduce. Protein Structure Graphs.
From proteopedia.org
Four levels of protein structure Proteopedia, life in 3D Protein Structure Graphs we obtain amino acid residue embeddings in protein structure through graph representation learning, utilize the. It is a database of protein structure topologies modeled as undirected, labeled graphs. in this short review paper, we survey studies that use graph learning techniques on proteins, and examine their. ptgl stands for protein topology graph library. here, we introduce. Protein Structure Graphs.
From microbenotes.com
Types of Protein Structure with Diagrams Protein Structure Graphs It is a database of protein structure topologies modeled as undirected, labeled graphs. we obtain amino acid residue embeddings in protein structure through graph representation learning, utilize the. here, we introduce deepfri, a graph convolutional network for predicting protein functions by leveraging sequence. ptgl stands for protein topology graph library. in this short review paper, we. Protein Structure Graphs.
From www.dreamstime.com
Protein Structure Levels from Amino Acid To Complex Molecule Outline Protein Structure Graphs ptgl stands for protein topology graph library. here, we introduce deepfri, a graph convolutional network for predicting protein functions by leveraging sequence. It is a database of protein structure topologies modeled as undirected, labeled graphs. in this short review paper, we survey studies that use graph learning techniques on proteins, and examine their. we obtain amino. Protein Structure Graphs.
From www.researchgate.net
RMSD graph of protein and proteinligand complex during 100 ns MD Protein Structure Graphs in this short review paper, we survey studies that use graph learning techniques on proteins, and examine their. we obtain amino acid residue embeddings in protein structure through graph representation learning, utilize the. ptgl stands for protein topology graph library. here, we introduce deepfri, a graph convolutional network for predicting protein functions by leveraging sequence. It. Protein Structure Graphs.
From thebiologynotes.com
Amino Acids and Proteins Structure, Types, Functions Protein Structure Graphs here, we introduce deepfri, a graph convolutional network for predicting protein functions by leveraging sequence. It is a database of protein structure topologies modeled as undirected, labeled graphs. ptgl stands for protein topology graph library. in this short review paper, we survey studies that use graph learning techniques on proteins, and examine their. we obtain amino. Protein Structure Graphs.
From bio.libretexts.org
3.7 Proteins Biology LibreTexts Protein Structure Graphs we obtain amino acid residue embeddings in protein structure through graph representation learning, utilize the. in this short review paper, we survey studies that use graph learning techniques on proteins, and examine their. ptgl stands for protein topology graph library. here, we introduce deepfri, a graph convolutional network for predicting protein functions by leveraging sequence. It. Protein Structure Graphs.
From www.medschoolcoach.com
Levels of Protein Structure MCAT Biochemistry MedSchoolCoach Protein Structure Graphs here, we introduce deepfri, a graph convolutional network for predicting protein functions by leveraging sequence. It is a database of protein structure topologies modeled as undirected, labeled graphs. ptgl stands for protein topology graph library. in this short review paper, we survey studies that use graph learning techniques on proteins, and examine their. we obtain amino. Protein Structure Graphs.
From typeset.io
(PDF) Graphein a Python Library for Geometric Deep Learning and Protein Structure Graphs we obtain amino acid residue embeddings in protein structure through graph representation learning, utilize the. in this short review paper, we survey studies that use graph learning techniques on proteins, and examine their. It is a database of protein structure topologies modeled as undirected, labeled graphs. ptgl stands for protein topology graph library. here, we introduce. Protein Structure Graphs.
From becksteinlab.physics.asu.edu
Identify secondary structure and topology — Practical 10 Protein Protein Structure Graphs we obtain amino acid residue embeddings in protein structure through graph representation learning, utilize the. in this short review paper, we survey studies that use graph learning techniques on proteins, and examine their. It is a database of protein structure topologies modeled as undirected, labeled graphs. ptgl stands for protein topology graph library. here, we introduce. Protein Structure Graphs.
From www.researchgate.net
Graph representation of the proteinligand complex. (a) 3D structure of Protein Structure Graphs in this short review paper, we survey studies that use graph learning techniques on proteins, and examine their. It is a database of protein structure topologies modeled as undirected, labeled graphs. ptgl stands for protein topology graph library. here, we introduce deepfri, a graph convolutional network for predicting protein functions by leveraging sequence. we obtain amino. Protein Structure Graphs.
From www.vectorstock.com
Levels protein structure from amino acids Vector Image Protein Structure Graphs ptgl stands for protein topology graph library. here, we introduce deepfri, a graph convolutional network for predicting protein functions by leveraging sequence. in this short review paper, we survey studies that use graph learning techniques on proteins, and examine their. we obtain amino acid residue embeddings in protein structure through graph representation learning, utilize the. It. Protein Structure Graphs.
From www.researchgate.net
Representing Protein Complexes as Graphs (A) Each protein complex is Protein Structure Graphs in this short review paper, we survey studies that use graph learning techniques on proteins, and examine their. ptgl stands for protein topology graph library. we obtain amino acid residue embeddings in protein structure through graph representation learning, utilize the. It is a database of protein structure topologies modeled as undirected, labeled graphs. here, we introduce. Protein Structure Graphs.
From www.researchgate.net
Typical examples of protein structures and the corresponding graph Protein Structure Graphs It is a database of protein structure topologies modeled as undirected, labeled graphs. here, we introduce deepfri, a graph convolutional network for predicting protein functions by leveraging sequence. in this short review paper, we survey studies that use graph learning techniques on proteins, and examine their. we obtain amino acid residue embeddings in protein structure through graph. Protein Structure Graphs.
From geneticeducation.co.in
What are the levels of Protein Structure? Protein Structure Graphs It is a database of protein structure topologies modeled as undirected, labeled graphs. we obtain amino acid residue embeddings in protein structure through graph representation learning, utilize the. in this short review paper, we survey studies that use graph learning techniques on proteins, and examine their. here, we introduce deepfri, a graph convolutional network for predicting protein. Protein Structure Graphs.
From deepai.org
PGR A Graph Repository of Protein 3DStructures DeepAI Protein Structure Graphs ptgl stands for protein topology graph library. we obtain amino acid residue embeddings in protein structure through graph representation learning, utilize the. It is a database of protein structure topologies modeled as undirected, labeled graphs. in this short review paper, we survey studies that use graph learning techniques on proteins, and examine their. here, we introduce. Protein Structure Graphs.
From analyticsdrift.com
Top 5 protein folding models Analytics Drift Data Science Protein Structure Graphs ptgl stands for protein topology graph library. here, we introduce deepfri, a graph convolutional network for predicting protein functions by leveraging sequence. we obtain amino acid residue embeddings in protein structure through graph representation learning, utilize the. It is a database of protein structure topologies modeled as undirected, labeled graphs. in this short review paper, we. Protein Structure Graphs.
From www.researchgate.net
Variety of protein structures. Examples of different types of proteins Protein Structure Graphs we obtain amino acid residue embeddings in protein structure through graph representation learning, utilize the. ptgl stands for protein topology graph library. It is a database of protein structure topologies modeled as undirected, labeled graphs. here, we introduce deepfri, a graph convolutional network for predicting protein functions by leveraging sequence. in this short review paper, we. Protein Structure Graphs.
From learningzoneflotum7j.z4.web.core.windows.net
Basics Of Protein Structure Protein Structure Graphs It is a database of protein structure topologies modeled as undirected, labeled graphs. in this short review paper, we survey studies that use graph learning techniques on proteins, and examine their. ptgl stands for protein topology graph library. here, we introduce deepfri, a graph convolutional network for predicting protein functions by leveraging sequence. we obtain amino. Protein Structure Graphs.
From www.mun.ca
Four levels of Protein Structure Protein Structure Graphs we obtain amino acid residue embeddings in protein structure through graph representation learning, utilize the. It is a database of protein structure topologies modeled as undirected, labeled graphs. here, we introduce deepfri, a graph convolutional network for predicting protein functions by leveraging sequence. ptgl stands for protein topology graph library. in this short review paper, we. Protein Structure Graphs.
From www.animalia-life.club
Quaternary Structure Of Protein Diagram Protein Structure Graphs It is a database of protein structure topologies modeled as undirected, labeled graphs. here, we introduce deepfri, a graph convolutional network for predicting protein functions by leveraging sequence. ptgl stands for protein topology graph library. in this short review paper, we survey studies that use graph learning techniques on proteins, and examine their. we obtain amino. Protein Structure Graphs.
From www.researchgate.net
Circular dichroism (CD) spectra of polypeptides and proteins with some Protein Structure Graphs It is a database of protein structure topologies modeled as undirected, labeled graphs. in this short review paper, we survey studies that use graph learning techniques on proteins, and examine their. here, we introduce deepfri, a graph convolutional network for predicting protein functions by leveraging sequence. we obtain amino acid residue embeddings in protein structure through graph. Protein Structure Graphs.
From civilmint.com
Various Levels of Protein Structure Protein Structure Graphs here, we introduce deepfri, a graph convolutional network for predicting protein functions by leveraging sequence. ptgl stands for protein topology graph library. in this short review paper, we survey studies that use graph learning techniques on proteins, and examine their. It is a database of protein structure topologies modeled as undirected, labeled graphs. we obtain amino. Protein Structure Graphs.
From www.researchgate.net
Graph Structure Is Related to ProteinProtein Interactions and Protein Structure Graphs ptgl stands for protein topology graph library. It is a database of protein structure topologies modeled as undirected, labeled graphs. in this short review paper, we survey studies that use graph learning techniques on proteins, and examine their. we obtain amino acid residue embeddings in protein structure through graph representation learning, utilize the. here, we introduce. Protein Structure Graphs.
From www.drawittoknowit.com
MCAT Biology & Biochemistry Glossary Protein Structure Class 1 Protein Structure Graphs ptgl stands for protein topology graph library. we obtain amino acid residue embeddings in protein structure through graph representation learning, utilize the. It is a database of protein structure topologies modeled as undirected, labeled graphs. in this short review paper, we survey studies that use graph learning techniques on proteins, and examine their. here, we introduce. Protein Structure Graphs.
From www.semanticscholar.org
Figure 1 from Protein Structure and Sequence Generation with Protein Structure Graphs here, we introduce deepfri, a graph convolutional network for predicting protein functions by leveraging sequence. in this short review paper, we survey studies that use graph learning techniques on proteins, and examine their. we obtain amino acid residue embeddings in protein structure through graph representation learning, utilize the. It is a database of protein structure topologies modeled. Protein Structure Graphs.
From www.visual-computing.org
Making group structures visible Visual Computing BLOG Protein Structure Graphs in this short review paper, we survey studies that use graph learning techniques on proteins, and examine their. we obtain amino acid residue embeddings in protein structure through graph representation learning, utilize the. It is a database of protein structure topologies modeled as undirected, labeled graphs. ptgl stands for protein topology graph library. here, we introduce. Protein Structure Graphs.
From www.researchgate.net
Proteinprotein interaction graph. Download Scientific Diagram Protein Structure Graphs in this short review paper, we survey studies that use graph learning techniques on proteins, and examine their. It is a database of protein structure topologies modeled as undirected, labeled graphs. ptgl stands for protein topology graph library. here, we introduce deepfri, a graph convolutional network for predicting protein functions by leveraging sequence. we obtain amino. Protein Structure Graphs.
From www.researchgate.net
Protein topology graphs based on PDB structures constructed using the Protein Structure Graphs It is a database of protein structure topologies modeled as undirected, labeled graphs. here, we introduce deepfri, a graph convolutional network for predicting protein functions by leveraging sequence. we obtain amino acid residue embeddings in protein structure through graph representation learning, utilize the. ptgl stands for protein topology graph library. in this short review paper, we. Protein Structure Graphs.
From learningkrausovel4.z21.web.core.windows.net
Structure Of Proteins Biology Protein Structure Graphs It is a database of protein structure topologies modeled as undirected, labeled graphs. in this short review paper, we survey studies that use graph learning techniques on proteins, and examine their. we obtain amino acid residue embeddings in protein structure through graph representation learning, utilize the. ptgl stands for protein topology graph library. here, we introduce. Protein Structure Graphs.
From www.researchgate.net
Protein sequence and structure alignment of JAK family PTK domain. (A Protein Structure Graphs It is a database of protein structure topologies modeled as undirected, labeled graphs. here, we introduce deepfri, a graph convolutional network for predicting protein functions by leveraging sequence. we obtain amino acid residue embeddings in protein structure through graph representation learning, utilize the. in this short review paper, we survey studies that use graph learning techniques on. Protein Structure Graphs.
From pubs.acs.org
Geometric Interaction Graph Neural Network for Predicting Protein Protein Structure Graphs It is a database of protein structure topologies modeled as undirected, labeled graphs. in this short review paper, we survey studies that use graph learning techniques on proteins, and examine their. ptgl stands for protein topology graph library. we obtain amino acid residue embeddings in protein structure through graph representation learning, utilize the. here, we introduce. Protein Structure Graphs.