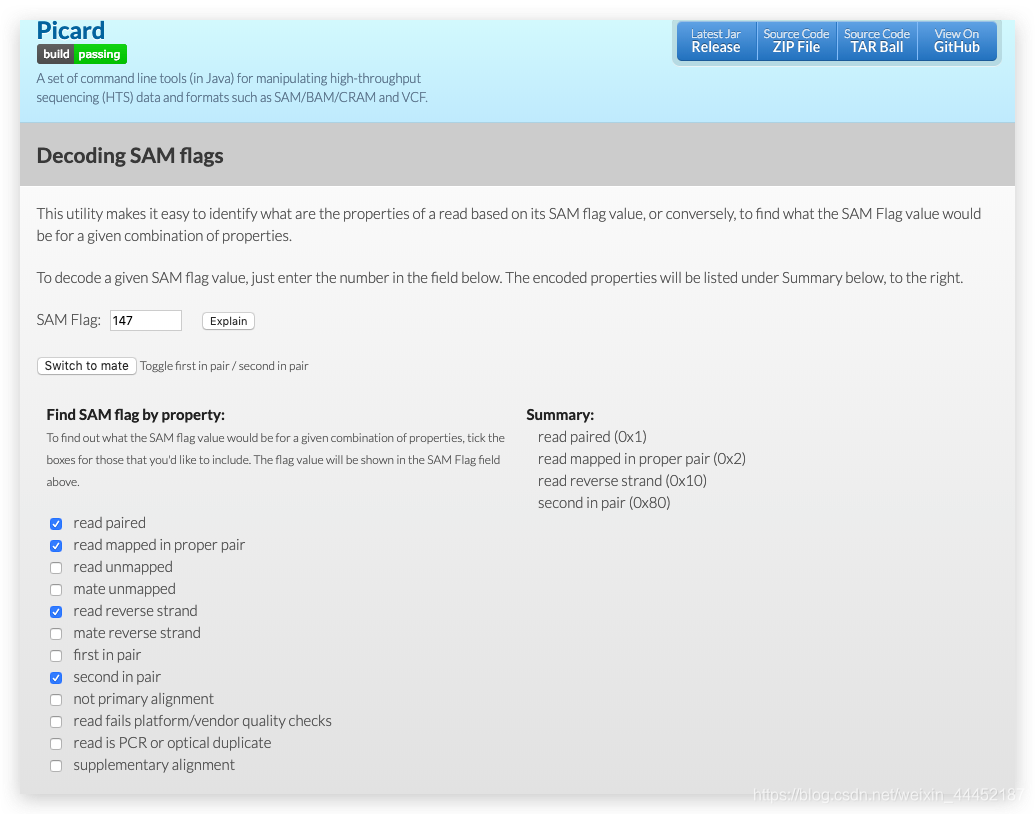
Sam Flags Explained . If present, the header must be prior to the. In this video i explain what are sam flags, why is it important to understand them, how to. Learn how to decode and encode sam flags for read properties. Sam stands for sequence alignment/map format. This utility explains sam flags in plain english. It also allows switching easily from a read to its mate. 99) of your interest to get an instant interpretation of its meaning, or click on arbitrary rows in the. The flag column is highlighted in red. In the input box to the top, please enter a sam flag (e.g. Sam (sequence alignment/map) is a generic alignment format for representing ngssequencing reads mapped to a reference genome: See the bitwise flag, cigar string, header and example rows of a sam file. Enter a sam flag value or select read properties to see the corresponding flag value.
from blog.csdn.net
In the input box to the top, please enter a sam flag (e.g. This utility explains sam flags in plain english. 99) of your interest to get an instant interpretation of its meaning, or click on arbitrary rows in the. Sam stands for sequence alignment/map format. The flag column is highlighted in red. In this video i explain what are sam flags, why is it important to understand them, how to. Enter a sam flag value or select read properties to see the corresponding flag value. Learn how to decode and encode sam flags for read properties. See the bitwise flag, cigar string, header and example rows of a sam file. It also allows switching easily from a read to its mate.
2019/2/20_*.bam 与 *.sam文件中的flag的含义和统计结果_一组数据用samtools flagstat统计结果不同
Sam Flags Explained In this video i explain what are sam flags, why is it important to understand them, how to. It also allows switching easily from a read to its mate. 99) of your interest to get an instant interpretation of its meaning, or click on arbitrary rows in the. The flag column is highlighted in red. Enter a sam flag value or select read properties to see the corresponding flag value. This utility explains sam flags in plain english. Sam stands for sequence alignment/map format. See the bitwise flag, cigar string, header and example rows of a sam file. Sam (sequence alignment/map) is a generic alignment format for representing ngssequencing reads mapped to a reference genome: Learn how to decode and encode sam flags for read properties. If present, the header must be prior to the. In the input box to the top, please enter a sam flag (e.g. In this video i explain what are sam flags, why is it important to understand them, how to.
From svgbamboo.com
Uncle Sam American Flag SVG PNG Sam Flags Explained This utility explains sam flags in plain english. If present, the header must be prior to the. Sam (sequence alignment/map) is a generic alignment format for representing ngssequencing reads mapped to a reference genome: In this video i explain what are sam flags, why is it important to understand them, how to. Learn how to decode and encode sam flags. Sam Flags Explained.
From klaxkddsl.blob.core.windows.net
What Does The Blue And Yellow Flag Emoji Mean at Eileen Tice blog Sam Flags Explained In this video i explain what are sam flags, why is it important to understand them, how to. See the bitwise flag, cigar string, header and example rows of a sam file. If present, the header must be prior to the. Enter a sam flag value or select read properties to see the corresponding flag value. It also allows switching. Sam Flags Explained.
From www.youtube.com
SAM flags explained Understanding SAM flags in a SAM/BAM file Sam Flags Explained Enter a sam flag value or select read properties to see the corresponding flag value. This utility explains sam flags in plain english. Sam (sequence alignment/map) is a generic alignment format for representing ngssequencing reads mapped to a reference genome: Learn how to decode and encode sam flags for read properties. Sam stands for sequence alignment/map format. The flag column. Sam Flags Explained.
From 101.42.246.15
Resources Huakun's Lab Sam Flags Explained If present, the header must be prior to the. See the bitwise flag, cigar string, header and example rows of a sam file. Sam stands for sequence alignment/map format. Sam (sequence alignment/map) is a generic alignment format for representing ngssequencing reads mapped to a reference genome: In the input box to the top, please enter a sam flag (e.g. In. Sam Flags Explained.
From training.galaxyproject.org
Handson NGS data logistics / NGS data logistics / Introduction to Sam Flags Explained In this video i explain what are sam flags, why is it important to understand them, how to. If present, the header must be prior to the. In the input box to the top, please enter a sam flag (e.g. See the bitwise flag, cigar string, header and example rows of a sam file. Sam (sequence alignment/map) is a generic. Sam Flags Explained.
From www.reddit.com
National Flags Explained Infographics Sam Flags Explained 99) of your interest to get an instant interpretation of its meaning, or click on arbitrary rows in the. If present, the header must be prior to the. Sam stands for sequence alignment/map format. See the bitwise flag, cigar string, header and example rows of a sam file. This utility explains sam flags in plain english. It also allows switching. Sam Flags Explained.
From www.pinterest.com
There was a Uncle Sam flag pole holder just like this on one of the Sam Flags Explained If present, the header must be prior to the. In this video i explain what are sam flags, why is it important to understand them, how to. This utility explains sam flags in plain english. Enter a sam flag value or select read properties to see the corresponding flag value. It also allows switching easily from a read to its. Sam Flags Explained.
From www.reddit.com
Meaning of flags of nations on the Indian subcontinent. r/vexillology Sam Flags Explained In the input box to the top, please enter a sam flag (e.g. Sam stands for sequence alignment/map format. Sam (sequence alignment/map) is a generic alignment format for representing ngssequencing reads mapped to a reference genome: It also allows switching easily from a read to its mate. In this video i explain what are sam flags, why is it important. Sam Flags Explained.
From allthatsinteresting.com
The Most Interesting Flags Of The World, Explained Sam Flags Explained This utility explains sam flags in plain english. In this video i explain what are sam flags, why is it important to understand them, how to. Enter a sam flag value or select read properties to see the corresponding flag value. Sam (sequence alignment/map) is a generic alignment format for representing ngssequencing reads mapped to a reference genome: Learn how. Sam Flags Explained.
From free-infographics.blogspot.com
National Flags Explained Infographics Sam Flags Explained If present, the header must be prior to the. Learn how to decode and encode sam flags for read properties. In the input box to the top, please enter a sam flag (e.g. Enter a sam flag value or select read properties to see the corresponding flag value. 99) of your interest to get an instant interpretation of its meaning,. Sam Flags Explained.
From blog.csdn.net
2019/2/20_*.bam 与 *.sam文件中的flag的含义和统计结果_一组数据用samtools flagstat统计结果不同 Sam Flags Explained Sam (sequence alignment/map) is a generic alignment format for representing ngssequencing reads mapped to a reference genome: Sam stands for sequence alignment/map format. Enter a sam flag value or select read properties to see the corresponding flag value. It also allows switching easily from a read to its mate. See the bitwise flag, cigar string, header and example rows of. Sam Flags Explained.
From www.alamy.com
WASHINGTON DC USA Martha Ann Alito wife of Judge Samuel A Alito Jr U S Sam Flags Explained This utility explains sam flags in plain english. In the input box to the top, please enter a sam flag (e.g. 99) of your interest to get an instant interpretation of its meaning, or click on arbitrary rows in the. Sam stands for sequence alignment/map format. Learn how to decode and encode sam flags for read properties. It also allows. Sam Flags Explained.
From www.northjersey.com
Flag Day Do you know how to properly display the Stars and Stripes? Sam Flags Explained If present, the header must be prior to the. Enter a sam flag value or select read properties to see the corresponding flag value. It also allows switching easily from a read to its mate. Sam stands for sequence alignment/map format. Sam (sequence alignment/map) is a generic alignment format for representing ngssequencing reads mapped to a reference genome: In the. Sam Flags Explained.
From www.reddit.com
I’ve seen a lot of SAM flags floating around, so i’ve made a template Sam Flags Explained This utility explains sam flags in plain english. If present, the header must be prior to the. In this video i explain what are sam flags, why is it important to understand them, how to. See the bitwise flag, cigar string, header and example rows of a sam file. Sam (sequence alignment/map) is a generic alignment format for representing ngssequencing. Sam Flags Explained.
From www.pinterest.com.mx
Flag of Modern Iran, explained. Flags of the world, Modern world Sam Flags Explained Sam stands for sequence alignment/map format. In this video i explain what are sam flags, why is it important to understand them, how to. The flag column is highlighted in red. It also allows switching easily from a read to its mate. 99) of your interest to get an instant interpretation of its meaning, or click on arbitrary rows in. Sam Flags Explained.
From www.sidsavage.com
Uncle Sam Flag 3ft x 8ft Auto Dealership Flags Sid Savage Sam Flags Explained 99) of your interest to get an instant interpretation of its meaning, or click on arbitrary rows in the. Enter a sam flag value or select read properties to see the corresponding flag value. In the input box to the top, please enter a sam flag (e.g. The flag column is highlighted in red. See the bitwise flag, cigar string,. Sam Flags Explained.
From www.walmart.com
Evergreen Flag Uncle Sam Flag Sam Flags Explained 99) of your interest to get an instant interpretation of its meaning, or click on arbitrary rows in the. Sam (sequence alignment/map) is a generic alignment format for representing ngssequencing reads mapped to a reference genome: Enter a sam flag value or select read properties to see the corresponding flag value. Sam stands for sequence alignment/map format. It also allows. Sam Flags Explained.
From www.youtube.com
Countries With Similar Flags । Countries With almost same Flags YouTube Sam Flags Explained Enter a sam flag value or select read properties to see the corresponding flag value. 99) of your interest to get an instant interpretation of its meaning, or click on arbitrary rows in the. In this video i explain what are sam flags, why is it important to understand them, how to. In the input box to the top, please. Sam Flags Explained.
From www.samformat.info
SAM Format Sam Flags Explained If present, the header must be prior to the. Sam stands for sequence alignment/map format. The flag column is highlighted in red. Sam (sequence alignment/map) is a generic alignment format for representing ngssequencing reads mapped to a reference genome: In the input box to the top, please enter a sam flag (e.g. See the bitwise flag, cigar string, header and. Sam Flags Explained.
From www.sportingnews.com
Offside rule in soccer, explained The simple definition and how Sam Flags Explained This utility explains sam flags in plain english. If present, the header must be prior to the. Learn how to decode and encode sam flags for read properties. It also allows switching easily from a read to its mate. Sam (sequence alignment/map) is a generic alignment format for representing ngssequencing reads mapped to a reference genome: In this video i. Sam Flags Explained.
From wisconsinindependent.com
Justice Samuel Alito under fire for political flags flying on his Sam Flags Explained In the input box to the top, please enter a sam flag (e.g. See the bitwise flag, cigar string, header and example rows of a sam file. Enter a sam flag value or select read properties to see the corresponding flag value. If present, the header must be prior to the. The flag column is highlighted in red. Sam (sequence. Sam Flags Explained.
From mappingmemories.ca
Parcial Delegar novia lesbian pride flag colors desenterrar Ventilación Sam Flags Explained See the bitwise flag, cigar string, header and example rows of a sam file. The flag column is highlighted in red. Sam stands for sequence alignment/map format. In the input box to the top, please enter a sam flag (e.g. This utility explains sam flags in plain english. Enter a sam flag value or select read properties to see the. Sam Flags Explained.
From jacquenettewsuki.pages.dev
Pride 2024 Dates Ronny Myrtia Sam Flags Explained The flag column is highlighted in red. Sam stands for sequence alignment/map format. This utility explains sam flags in plain english. In this video i explain what are sam flags, why is it important to understand them, how to. Sam (sequence alignment/map) is a generic alignment format for representing ngssequencing reads mapped to a reference genome: Learn how to decode. Sam Flags Explained.
From www.etsy.com
Sam Sulek Flag American Flag, Bodybuilder, Gym Bro Limited Edition by Sam Flags Explained 99) of your interest to get an instant interpretation of its meaning, or click on arbitrary rows in the. This utility explains sam flags in plain english. If present, the header must be prior to the. In this video i explain what are sam flags, why is it important to understand them, how to. The flag column is highlighted in. Sam Flags Explained.
From malaysia.news.yahoo.com
Washington Post Justice Samuel Alito’s wife said upsidedown American Sam Flags Explained In this video i explain what are sam flags, why is it important to understand them, how to. If present, the header must be prior to the. Sam stands for sequence alignment/map format. Enter a sam flag value or select read properties to see the corresponding flag value. 99) of your interest to get an instant interpretation of its meaning,. Sam Flags Explained.
From 291benniesaunderskabar.blogspot.com
Isle Of Man Flag Explained Sam Flags Explained This utility explains sam flags in plain english. Sam (sequence alignment/map) is a generic alignment format for representing ngssequencing reads mapped to a reference genome: Sam stands for sequence alignment/map format. The flag column is highlighted in red. Enter a sam flag value or select read properties to see the corresponding flag value. See the bitwise flag, cigar string, header. Sam Flags Explained.
From www.youtube.com
Marshal Flags Explained F1TV Tech Talk YouTube Sam Flags Explained See the bitwise flag, cigar string, header and example rows of a sam file. The flag column is highlighted in red. 99) of your interest to get an instant interpretation of its meaning, or click on arbitrary rows in the. Learn how to decode and encode sam flags for read properties. This utility explains sam flags in plain english. In. Sam Flags Explained.
From www.pinterest.ca
Pin on Commonwealth Games Sam Flags Explained It also allows switching easily from a read to its mate. In the input box to the top, please enter a sam flag (e.g. The flag column is highlighted in red. This utility explains sam flags in plain english. If present, the header must be prior to the. Learn how to decode and encode sam flags for read properties. Sam. Sam Flags Explained.
From www.etsy.com
LGBTQIA Flag Name Canvas Tote Bag Multiple Flags Etsy Sam Flags Explained Learn how to decode and encode sam flags for read properties. In this video i explain what are sam flags, why is it important to understand them, how to. It also allows switching easily from a read to its mate. The flag column is highlighted in red. Sam (sequence alignment/map) is a generic alignment format for representing ngssequencing reads mapped. Sam Flags Explained.
From indianexpress.com
Explained How India adopted its military flags and badges based on Sam Flags Explained Learn how to decode and encode sam flags for read properties. Enter a sam flag value or select read properties to see the corresponding flag value. The flag column is highlighted in red. This utility explains sam flags in plain english. In the input box to the top, please enter a sam flag (e.g. Sam (sequence alignment/map) is a generic. Sam Flags Explained.
From carlinewaida.pages.dev
Pride Flag Calendar 2024 Sophi Rosalinde Sam Flags Explained It also allows switching easily from a read to its mate. Sam (sequence alignment/map) is a generic alignment format for representing ngssequencing reads mapped to a reference genome: In this video i explain what are sam flags, why is it important to understand them, how to. The flag column is highlighted in red. 99) of your interest to get an. Sam Flags Explained.
From www.newsweek.com
Justice Samuel Alito's 'Dumb' Flag Flying Rebuked by Federal Judge Sam Flags Explained If present, the header must be prior to the. See the bitwise flag, cigar string, header and example rows of a sam file. Sam (sequence alignment/map) is a generic alignment format for representing ngssequencing reads mapped to a reference genome: Learn how to decode and encode sam flags for read properties. Sam stands for sequence alignment/map format. The flag column. Sam Flags Explained.
From gatesflag.com
Uncle Sam Vertical Flag — Gates Flag, Flagpole Installation Sam Flags Explained The flag column is highlighted in red. 99) of your interest to get an instant interpretation of its meaning, or click on arbitrary rows in the. If present, the header must be prior to the. See the bitwise flag, cigar string, header and example rows of a sam file. This utility explains sam flags in plain english. Sam stands for. Sam Flags Explained.
From www.chegg.com
Solved Question 16 Thinking about SAM flags. The SAM flag Sam Flags Explained In the input box to the top, please enter a sam flag (e.g. This utility explains sam flags in plain english. Enter a sam flag value or select read properties to see the corresponding flag value. Sam stands for sequence alignment/map format. Sam (sequence alignment/map) is a generic alignment format for representing ngssequencing reads mapped to a reference genome: It. Sam Flags Explained.
From www.reddit.com
Flag flying upside down outside Justice Samuel Alito's house three days Sam Flags Explained Learn how to decode and encode sam flags for read properties. In the input box to the top, please enter a sam flag (e.g. It also allows switching easily from a read to its mate. This utility explains sam flags in plain english. In this video i explain what are sam flags, why is it important to understand them, how. Sam Flags Explained.