Protein Binding Affinity Measurement . A database of measured binding affinities, focusing chiefly on the interactions of proteiproteinsdered to be drug. We show that probound quantifies transcription factor (tf) behavior with models that predict binding affinity over a range exceeding. The binding affinity (k a) and the quantity c k a [protein] are shown in the panels. In the left chart for la, the. In this study, we have developed a novel experimental procedure using microscale thermophoresis (mst) directly from cell. We evaluated the potential for new features to improve affinity prediction models by calculating the pearson correlation between. For c > 1000, only a lower limit for the binding affinity can be.
from www.rapidnovor.com
A database of measured binding affinities, focusing chiefly on the interactions of proteiproteinsdered to be drug. In the left chart for la, the. We show that probound quantifies transcription factor (tf) behavior with models that predict binding affinity over a range exceeding. We evaluated the potential for new features to improve affinity prediction models by calculating the pearson correlation between. For c > 1000, only a lower limit for the binding affinity can be. The binding affinity (k a) and the quantity c k a [protein] are shown in the panels. In this study, we have developed a novel experimental procedure using microscale thermophoresis (mst) directly from cell.
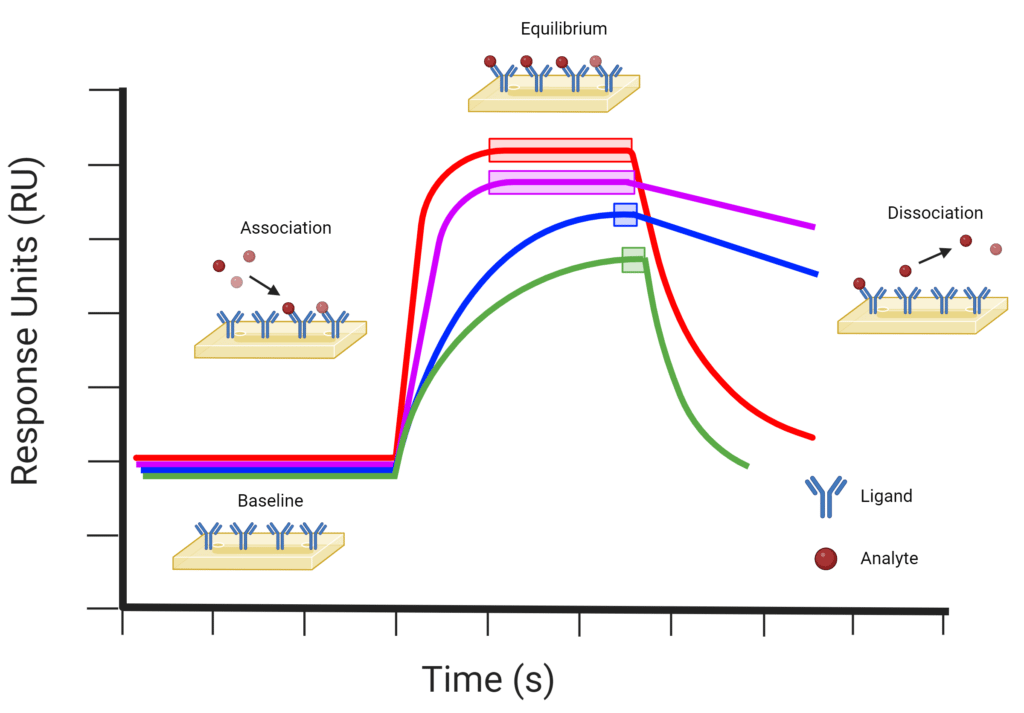
Antibody Affinity, Avidity Single vs Multivalent Interaction
Protein Binding Affinity Measurement In this study, we have developed a novel experimental procedure using microscale thermophoresis (mst) directly from cell. A database of measured binding affinities, focusing chiefly on the interactions of proteiproteinsdered to be drug. The binding affinity (k a) and the quantity c k a [protein] are shown in the panels. For c > 1000, only a lower limit for the binding affinity can be. We show that probound quantifies transcription factor (tf) behavior with models that predict binding affinity over a range exceeding. In the left chart for la, the. We evaluated the potential for new features to improve affinity prediction models by calculating the pearson correlation between. In this study, we have developed a novel experimental procedure using microscale thermophoresis (mst) directly from cell.
From www.frontiersin.org
Frontiers Machine learning methods for proteinprotein binding Protein Binding Affinity Measurement The binding affinity (k a) and the quantity c k a [protein] are shown in the panels. We show that probound quantifies transcription factor (tf) behavior with models that predict binding affinity over a range exceeding. In this study, we have developed a novel experimental procedure using microscale thermophoresis (mst) directly from cell. A database of measured binding affinities, focusing. Protein Binding Affinity Measurement.
From www.researchgate.net
Biacore measurement of the binding affinity of monoclonal 1C6. (A Protein Binding Affinity Measurement We show that probound quantifies transcription factor (tf) behavior with models that predict binding affinity over a range exceeding. In the left chart for la, the. We evaluated the potential for new features to improve affinity prediction models by calculating the pearson correlation between. A database of measured binding affinities, focusing chiefly on the interactions of proteiproteinsdered to be drug.. Protein Binding Affinity Measurement.
From www.researchgate.net
Proteinprotein binding affinities and variant evolution prediction Protein Binding Affinity Measurement For c > 1000, only a lower limit for the binding affinity can be. We show that probound quantifies transcription factor (tf) behavior with models that predict binding affinity over a range exceeding. In this study, we have developed a novel experimental procedure using microscale thermophoresis (mst) directly from cell. In the left chart for la, the. We evaluated the. Protein Binding Affinity Measurement.
From pubs.acs.org
Protein Binding Affinity of Polymeric Nanoparticles as a Direct Protein Binding Affinity Measurement For c > 1000, only a lower limit for the binding affinity can be. In the left chart for la, the. The binding affinity (k a) and the quantity c k a [protein] are shown in the panels. In this study, we have developed a novel experimental procedure using microscale thermophoresis (mst) directly from cell. We show that probound quantifies. Protein Binding Affinity Measurement.
From elifesciences.org
How to measure and evaluate binding affinities eLife Protein Binding Affinity Measurement In the left chart for la, the. For c > 1000, only a lower limit for the binding affinity can be. We show that probound quantifies transcription factor (tf) behavior with models that predict binding affinity over a range exceeding. The binding affinity (k a) and the quantity c k a [protein] are shown in the panels. In this study,. Protein Binding Affinity Measurement.
From www.researchgate.net
PBMdetermined proteinDNA binding affinities (Kd's) and comparison Protein Binding Affinity Measurement The binding affinity (k a) and the quantity c k a [protein] are shown in the panels. We evaluated the potential for new features to improve affinity prediction models by calculating the pearson correlation between. For c > 1000, only a lower limit for the binding affinity can be. In the left chart for la, the. We show that probound. Protein Binding Affinity Measurement.
From www.researchgate.net
The binding affinity of BmILF to imotif structures. EMSA for the Protein Binding Affinity Measurement We show that probound quantifies transcription factor (tf) behavior with models that predict binding affinity over a range exceeding. In this study, we have developed a novel experimental procedure using microscale thermophoresis (mst) directly from cell. A database of measured binding affinities, focusing chiefly on the interactions of proteiproteinsdered to be drug. In the left chart for la, the. For. Protein Binding Affinity Measurement.
From www.doccheck.com
Binding Affinity Measurement DocCheck Protein Binding Affinity Measurement In the left chart for la, the. In this study, we have developed a novel experimental procedure using microscale thermophoresis (mst) directly from cell. A database of measured binding affinities, focusing chiefly on the interactions of proteiproteinsdered to be drug. The binding affinity (k a) and the quantity c k a [protein] are shown in the panels. We evaluated the. Protein Binding Affinity Measurement.
From www.researchgate.net
Measurement of binding affinities among the S. cerevisiae GLFG Protein Binding Affinity Measurement In the left chart for la, the. We show that probound quantifies transcription factor (tf) behavior with models that predict binding affinity over a range exceeding. The binding affinity (k a) and the quantity c k a [protein] are shown in the panels. A database of measured binding affinities, focusing chiefly on the interactions of proteiproteinsdered to be drug. For. Protein Binding Affinity Measurement.
From 2bind.com
Aptamers for proteins Measuring binding affinity • 2bind MST Service Protein Binding Affinity Measurement In this study, we have developed a novel experimental procedure using microscale thermophoresis (mst) directly from cell. The binding affinity (k a) and the quantity c k a [protein] are shown in the panels. In the left chart for la, the. For c > 1000, only a lower limit for the binding affinity can be. We evaluated the potential for. Protein Binding Affinity Measurement.
From elifesciences.org
How to measure and evaluate binding affinities eLife Protein Binding Affinity Measurement A database of measured binding affinities, focusing chiefly on the interactions of proteiproteinsdered to be drug. The binding affinity (k a) and the quantity c k a [protein] are shown in the panels. In the left chart for la, the. We show that probound quantifies transcription factor (tf) behavior with models that predict binding affinity over a range exceeding. For. Protein Binding Affinity Measurement.
From www.rapidnovor.com
Antibody Affinity, Avidity Single vs Multivalent Interaction Protein Binding Affinity Measurement In this study, we have developed a novel experimental procedure using microscale thermophoresis (mst) directly from cell. In the left chart for la, the. We show that probound quantifies transcription factor (tf) behavior with models that predict binding affinity over a range exceeding. We evaluated the potential for new features to improve affinity prediction models by calculating the pearson correlation. Protein Binding Affinity Measurement.
From elifesciences.org
Figures and data in How to measure and evaluate binding affinities eLife Protein Binding Affinity Measurement We evaluated the potential for new features to improve affinity prediction models by calculating the pearson correlation between. The binding affinity (k a) and the quantity c k a [protein] are shown in the panels. For c > 1000, only a lower limit for the binding affinity can be. We show that probound quantifies transcription factor (tf) behavior with models. Protein Binding Affinity Measurement.
From pubs.acs.org
Measurement of In Vivo Protein Binding Affinities in a Signaling Protein Binding Affinity Measurement We evaluated the potential for new features to improve affinity prediction models by calculating the pearson correlation between. For c > 1000, only a lower limit for the binding affinity can be. A database of measured binding affinities, focusing chiefly on the interactions of proteiproteinsdered to be drug. The binding affinity (k a) and the quantity c k a [protein]. Protein Binding Affinity Measurement.
From www.researchgate.net
The affinity measurement and structural modeling between the mutated Protein Binding Affinity Measurement In the left chart for la, the. In this study, we have developed a novel experimental procedure using microscale thermophoresis (mst) directly from cell. We evaluated the potential for new features to improve affinity prediction models by calculating the pearson correlation between. We show that probound quantifies transcription factor (tf) behavior with models that predict binding affinity over a range. Protein Binding Affinity Measurement.
From www.semanticscholar.org
Table 1 from Measurement of In Vivo Protein Binding Affinities in a Protein Binding Affinity Measurement We evaluated the potential for new features to improve affinity prediction models by calculating the pearson correlation between. For c > 1000, only a lower limit for the binding affinity can be. In this study, we have developed a novel experimental procedure using microscale thermophoresis (mst) directly from cell. The binding affinity (k a) and the quantity c k a. Protein Binding Affinity Measurement.
From 2bind.com
Aptamers for proteins Measuring binding affinity • 2bind MST Service Protein Binding Affinity Measurement For c > 1000, only a lower limit for the binding affinity can be. In the left chart for la, the. In this study, we have developed a novel experimental procedure using microscale thermophoresis (mst) directly from cell. A database of measured binding affinities, focusing chiefly on the interactions of proteiproteinsdered to be drug. The binding affinity (k a) and. Protein Binding Affinity Measurement.
From 2bind.com
Aptamers for proteins Measuring binding affinity • 2bind MST Service Protein Binding Affinity Measurement In this study, we have developed a novel experimental procedure using microscale thermophoresis (mst) directly from cell. The binding affinity (k a) and the quantity c k a [protein] are shown in the panels. For c > 1000, only a lower limit for the binding affinity can be. In the left chart for la, the. We show that probound quantifies. Protein Binding Affinity Measurement.
From www.researchgate.net
Assessment of affinity of binding of the scFvhFcs to PTH1RECD by SPR Protein Binding Affinity Measurement In this study, we have developed a novel experimental procedure using microscale thermophoresis (mst) directly from cell. We evaluated the potential for new features to improve affinity prediction models by calculating the pearson correlation between. For c > 1000, only a lower limit for the binding affinity can be. We show that probound quantifies transcription factor (tf) behavior with models. Protein Binding Affinity Measurement.
From www.researchgate.net
Measurements of binding affinity and enzyme Measurements of Protein Binding Affinity Measurement In this study, we have developed a novel experimental procedure using microscale thermophoresis (mst) directly from cell. We show that probound quantifies transcription factor (tf) behavior with models that predict binding affinity over a range exceeding. The binding affinity (k a) and the quantity c k a [protein] are shown in the panels. For c > 1000, only a lower. Protein Binding Affinity Measurement.
From www.pnas.org
Correlation between the binding affinity and the conformational entropy Protein Binding Affinity Measurement In this study, we have developed a novel experimental procedure using microscale thermophoresis (mst) directly from cell. We show that probound quantifies transcription factor (tf) behavior with models that predict binding affinity over a range exceeding. We evaluated the potential for new features to improve affinity prediction models by calculating the pearson correlation between. A database of measured binding affinities,. Protein Binding Affinity Measurement.
From www.researchgate.net
DNA binding affinities of FOX proteins. (A) DNAbinding features of Protein Binding Affinity Measurement We evaluated the potential for new features to improve affinity prediction models by calculating the pearson correlation between. In the left chart for la, the. For c > 1000, only a lower limit for the binding affinity can be. The binding affinity (k a) and the quantity c k a [protein] are shown in the panels. A database of measured. Protein Binding Affinity Measurement.
From www.researchgate.net
Relative binding affinity of NISTmAb samples (A) Protein A binding Protein Binding Affinity Measurement We evaluated the potential for new features to improve affinity prediction models by calculating the pearson correlation between. In the left chart for la, the. We show that probound quantifies transcription factor (tf) behavior with models that predict binding affinity over a range exceeding. A database of measured binding affinities, focusing chiefly on the interactions of proteiproteinsdered to be drug.. Protein Binding Affinity Measurement.
From www.researchgate.net
Affinity maturation of HER2binding small proteins. (A) ELISA was used Protein Binding Affinity Measurement The binding affinity (k a) and the quantity c k a [protein] are shown in the panels. In this study, we have developed a novel experimental procedure using microscale thermophoresis (mst) directly from cell. In the left chart for la, the. A database of measured binding affinities, focusing chiefly on the interactions of proteiproteinsdered to be drug. We evaluated the. Protein Binding Affinity Measurement.
From www.neuromics.com
Protein Affinity Chromatography Protein Binding Affinity Measurement In this study, we have developed a novel experimental procedure using microscale thermophoresis (mst) directly from cell. We evaluated the potential for new features to improve affinity prediction models by calculating the pearson correlation between. A database of measured binding affinities, focusing chiefly on the interactions of proteiproteinsdered to be drug. We show that probound quantifies transcription factor (tf) behavior. Protein Binding Affinity Measurement.
From www.researchgate.net
Binding affinity measurements of Trastuzumab and Pertuzumab Protein Binding Affinity Measurement We evaluated the potential for new features to improve affinity prediction models by calculating the pearson correlation between. The binding affinity (k a) and the quantity c k a [protein] are shown in the panels. For c > 1000, only a lower limit for the binding affinity can be. In the left chart for la, the. In this study, we. Protein Binding Affinity Measurement.
From www.researchgate.net
(PDF) A robust assay to measure DNA topologydependent protein binding Protein Binding Affinity Measurement The binding affinity (k a) and the quantity c k a [protein] are shown in the panels. In this study, we have developed a novel experimental procedure using microscale thermophoresis (mst) directly from cell. For c > 1000, only a lower limit for the binding affinity can be. In the left chart for la, the. We show that probound quantifies. Protein Binding Affinity Measurement.
From www.researchgate.net
ITC measurement of the binding affinity between SARSCoV2 N and Ubl1 a Protein Binding Affinity Measurement A database of measured binding affinities, focusing chiefly on the interactions of proteiproteinsdered to be drug. We evaluated the potential for new features to improve affinity prediction models by calculating the pearson correlation between. We show that probound quantifies transcription factor (tf) behavior with models that predict binding affinity over a range exceeding. In the left chart for la, the.. Protein Binding Affinity Measurement.
From www.caframolabsolutions.com
Protein Affinity Chromatography Caframo Lab Solutions Protein Binding Affinity Measurement The binding affinity (k a) and the quantity c k a [protein] are shown in the panels. For c > 1000, only a lower limit for the binding affinity can be. In the left chart for la, the. We show that probound quantifies transcription factor (tf) behavior with models that predict binding affinity over a range exceeding. A database of. Protein Binding Affinity Measurement.
From www.researchgate.net
Binding affinity measurement using BLI. (A) The binding affinity of Protein Binding Affinity Measurement For c > 1000, only a lower limit for the binding affinity can be. We evaluated the potential for new features to improve affinity prediction models by calculating the pearson correlation between. We show that probound quantifies transcription factor (tf) behavior with models that predict binding affinity over a range exceeding. In the left chart for la, the. A database. Protein Binding Affinity Measurement.
From www.researchgate.net
Measurement of binding affinity by surface plasmon resonance (SPR). (A Protein Binding Affinity Measurement The binding affinity (k a) and the quantity c k a [protein] are shown in the panels. For c > 1000, only a lower limit for the binding affinity can be. In the left chart for la, the. We show that probound quantifies transcription factor (tf) behavior with models that predict binding affinity over a range exceeding. A database of. Protein Binding Affinity Measurement.
From elifesciences.org
How to measure and evaluate binding affinities eLife Protein Binding Affinity Measurement In this study, we have developed a novel experimental procedure using microscale thermophoresis (mst) directly from cell. We show that probound quantifies transcription factor (tf) behavior with models that predict binding affinity over a range exceeding. The binding affinity (k a) and the quantity c k a [protein] are shown in the panels. A database of measured binding affinities, focusing. Protein Binding Affinity Measurement.
From www.researchgate.net
Measurement of the binding affinities between the RBDs and ACE2 by SPR Protein Binding Affinity Measurement We evaluated the potential for new features to improve affinity prediction models by calculating the pearson correlation between. We show that probound quantifies transcription factor (tf) behavior with models that predict binding affinity over a range exceeding. A database of measured binding affinities, focusing chiefly on the interactions of proteiproteinsdered to be drug. In this study, we have developed a. Protein Binding Affinity Measurement.
From exobdajhi.blob.core.windows.net
Kd Binding Affinity Calculation at Durand blog Protein Binding Affinity Measurement In this study, we have developed a novel experimental procedure using microscale thermophoresis (mst) directly from cell. The binding affinity (k a) and the quantity c k a [protein] are shown in the panels. We evaluated the potential for new features to improve affinity prediction models by calculating the pearson correlation between. For c > 1000, only a lower limit. Protein Binding Affinity Measurement.
From www.researchgate.net
Measurement of the cdiGMP binding affinity of Cbp1 and Cbp1R320A by Protein Binding Affinity Measurement In this study, we have developed a novel experimental procedure using microscale thermophoresis (mst) directly from cell. A database of measured binding affinities, focusing chiefly on the interactions of proteiproteinsdered to be drug. The binding affinity (k a) and the quantity c k a [protein] are shown in the panels. We evaluated the potential for new features to improve affinity. Protein Binding Affinity Measurement.