Cufflinks Bioinformatics Tutorial . The paper includes a diagram (figure 2) describing how the various parts of the cufflinks package (and its companion tool tophat) fit together. #how does cufflinks calculate transcript abundances? “treated” vs “control”, “disease” vs “control”, “tissue a” vs “tissue b”. Cuffnorm takes a gtf2/gff3 file of transcripts as input, along with two or more sam, bam, or cxb files for two or more samples. This tutorial illustrates how to perform basic differential expression test between 2 groups of samples (for instance: #how does cufflinks assemble transcripts? The cufflinks suite includes a.
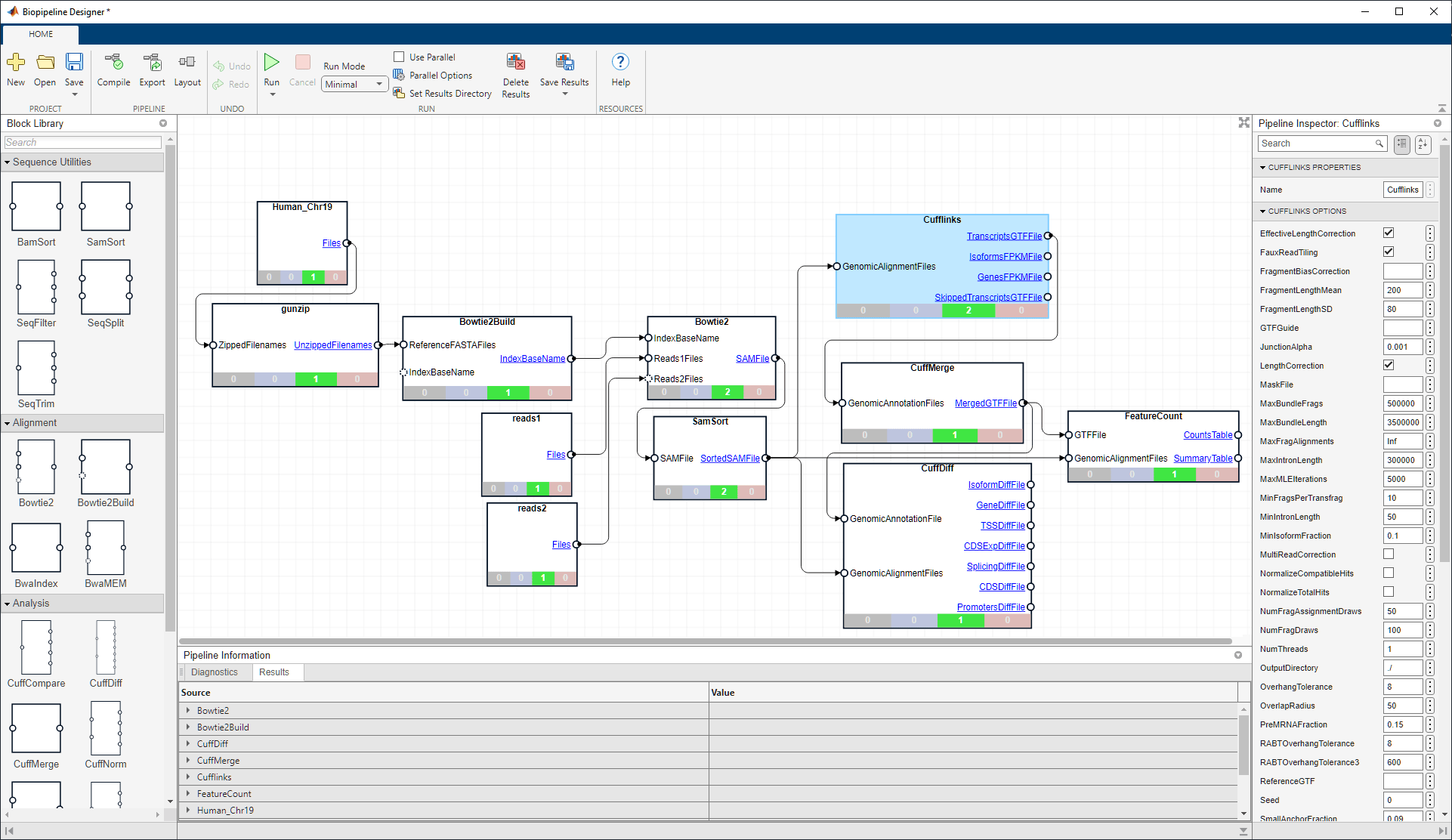
from ww2.mathworks.cn
#how does cufflinks assemble transcripts? The cufflinks suite includes a. The paper includes a diagram (figure 2) describing how the various parts of the cufflinks package (and its companion tool tophat) fit together. #how does cufflinks calculate transcript abundances? Cuffnorm takes a gtf2/gff3 file of transcripts as input, along with two or more sam, bam, or cxb files for two or more samples. This tutorial illustrates how to perform basic differential expression test between 2 groups of samples (for instance: “treated” vs “control”, “disease” vs “control”, “tissue a” vs “tissue b”.
Build and run bioinformatics pipelines MATLAB MathWorks 中国
Cufflinks Bioinformatics Tutorial #how does cufflinks calculate transcript abundances? The cufflinks suite includes a. The paper includes a diagram (figure 2) describing how the various parts of the cufflinks package (and its companion tool tophat) fit together. “treated” vs “control”, “disease” vs “control”, “tissue a” vs “tissue b”. #how does cufflinks assemble transcripts? #how does cufflinks calculate transcript abundances? This tutorial illustrates how to perform basic differential expression test between 2 groups of samples (for instance: Cuffnorm takes a gtf2/gff3 file of transcripts as input, along with two or more sam, bam, or cxb files for two or more samples.
From www.slideshare.net
Imgc2011 bioinformatics tutorial Cufflinks Bioinformatics Tutorial “treated” vs “control”, “disease” vs “control”, “tissue a” vs “tissue b”. The paper includes a diagram (figure 2) describing how the various parts of the cufflinks package (and its companion tool tophat) fit together. This tutorial illustrates how to perform basic differential expression test between 2 groups of samples (for instance: The cufflinks suite includes a. #how does cufflinks assemble. Cufflinks Bioinformatics Tutorial.
From www.embl.org
Bioinformatics tutorial EMBL ELLS Cufflinks Bioinformatics Tutorial “treated” vs “control”, “disease” vs “control”, “tissue a” vs “tissue b”. #how does cufflinks assemble transcripts? The cufflinks suite includes a. Cuffnorm takes a gtf2/gff3 file of transcripts as input, along with two or more sam, bam, or cxb files for two or more samples. This tutorial illustrates how to perform basic differential expression test between 2 groups of samples. Cufflinks Bioinformatics Tutorial.
From vectorartillustrationartworks.blogspot.com
tophat rna seq tutorial vectorartillustrationartworks Cufflinks Bioinformatics Tutorial The paper includes a diagram (figure 2) describing how the various parts of the cufflinks package (and its companion tool tophat) fit together. Cuffnorm takes a gtf2/gff3 file of transcripts as input, along with two or more sam, bam, or cxb files for two or more samples. This tutorial illustrates how to perform basic differential expression test between 2 groups. Cufflinks Bioinformatics Tutorial.
From www.youtube.com
5 Steps to Transitioning Into Bioinformatics As A Bio Student YouTube Cufflinks Bioinformatics Tutorial The cufflinks suite includes a. The paper includes a diagram (figure 2) describing how the various parts of the cufflinks package (and its companion tool tophat) fit together. This tutorial illustrates how to perform basic differential expression test between 2 groups of samples (for instance: Cuffnorm takes a gtf2/gff3 file of transcripts as input, along with two or more sam,. Cufflinks Bioinformatics Tutorial.
From www.youtube.com
Convert Ensembl IDs to Gene Symbols in R Biomart Mr. BioinformatiX Cufflinks Bioinformatics Tutorial “treated” vs “control”, “disease” vs “control”, “tissue a” vs “tissue b”. The paper includes a diagram (figure 2) describing how the various parts of the cufflinks package (and its companion tool tophat) fit together. The cufflinks suite includes a. #how does cufflinks calculate transcript abundances? #how does cufflinks assemble transcripts? Cuffnorm takes a gtf2/gff3 file of transcripts as input, along. Cufflinks Bioinformatics Tutorial.
From kevfo.github.io
BS4017 High Throughput Bioinformatics 6 Transcriptomes Experiment Cufflinks Bioinformatics Tutorial #how does cufflinks assemble transcripts? Cuffnorm takes a gtf2/gff3 file of transcripts as input, along with two or more sam, bam, or cxb files for two or more samples. #how does cufflinks calculate transcript abundances? This tutorial illustrates how to perform basic differential expression test between 2 groups of samples (for instance: The paper includes a diagram (figure 2) describing. Cufflinks Bioinformatics Tutorial.
From studylib.net
CMSC702_Cufflinks_Final Center for Bioinformatics and Cufflinks Bioinformatics Tutorial The cufflinks suite includes a. Cuffnorm takes a gtf2/gff3 file of transcripts as input, along with two or more sam, bam, or cxb files for two or more samples. #how does cufflinks calculate transcript abundances? This tutorial illustrates how to perform basic differential expression test between 2 groups of samples (for instance: “treated” vs “control”, “disease” vs “control”, “tissue a”. Cufflinks Bioinformatics Tutorial.
From www.youtube.com
Biopython Bioinformatics Tutorial Global and Local Alignments Mr Cufflinks Bioinformatics Tutorial The paper includes a diagram (figure 2) describing how the various parts of the cufflinks package (and its companion tool tophat) fit together. #how does cufflinks assemble transcripts? Cuffnorm takes a gtf2/gff3 file of transcripts as input, along with two or more sam, bam, or cxb files for two or more samples. #how does cufflinks calculate transcript abundances? “treated” vs. Cufflinks Bioinformatics Tutorial.
From github.com
GitHub A tutorial on how to Cufflinks Bioinformatics Tutorial The paper includes a diagram (figure 2) describing how the various parts of the cufflinks package (and its companion tool tophat) fit together. This tutorial illustrates how to perform basic differential expression test between 2 groups of samples (for instance: #how does cufflinks calculate transcript abundances? The cufflinks suite includes a. Cuffnorm takes a gtf2/gff3 file of transcripts as input,. Cufflinks Bioinformatics Tutorial.
From www.biocode.org.uk
Bioinformatics Training For Beginners BioCode Cufflinks Bioinformatics Tutorial The cufflinks suite includes a. “treated” vs “control”, “disease” vs “control”, “tissue a” vs “tissue b”. Cuffnorm takes a gtf2/gff3 file of transcripts as input, along with two or more sam, bam, or cxb files for two or more samples. The paper includes a diagram (figure 2) describing how the various parts of the cufflinks package (and its companion tool. Cufflinks Bioinformatics Tutorial.
From www.youtube.com
Galaxy Tutorials How to use Galaxy for Bioinformatics (Beginners Cufflinks Bioinformatics Tutorial The cufflinks suite includes a. #how does cufflinks assemble transcripts? Cuffnorm takes a gtf2/gff3 file of transcripts as input, along with two or more sam, bam, or cxb files for two or more samples. “treated” vs “control”, “disease” vs “control”, “tissue a” vs “tissue b”. #how does cufflinks calculate transcript abundances? This tutorial illustrates how to perform basic differential expression. Cufflinks Bioinformatics Tutorial.
From github.com
GitHub mathworks/cufflinks Cufflinks compiled binaries for the Cufflinks Bioinformatics Tutorial #how does cufflinks assemble transcripts? Cuffnorm takes a gtf2/gff3 file of transcripts as input, along with two or more sam, bam, or cxb files for two or more samples. “treated” vs “control”, “disease” vs “control”, “tissue a” vs “tissue b”. #how does cufflinks calculate transcript abundances? The cufflinks suite includes a. The paper includes a diagram (figure 2) describing how. Cufflinks Bioinformatics Tutorial.
From www.embl.org
Bioinformatics tutorial EMBL ELLS Cufflinks Bioinformatics Tutorial The paper includes a diagram (figure 2) describing how the various parts of the cufflinks package (and its companion tool tophat) fit together. #how does cufflinks assemble transcripts? #how does cufflinks calculate transcript abundances? This tutorial illustrates how to perform basic differential expression test between 2 groups of samples (for instance: “treated” vs “control”, “disease” vs “control”, “tissue a” vs. Cufflinks Bioinformatics Tutorial.
From www.learntoupgrade.com
How To Bioinformatics Scientist Cufflinks Bioinformatics Tutorial #how does cufflinks assemble transcripts? The cufflinks suite includes a. The paper includes a diagram (figure 2) describing how the various parts of the cufflinks package (and its companion tool tophat) fit together. #how does cufflinks calculate transcript abundances? Cuffnorm takes a gtf2/gff3 file of transcripts as input, along with two or more sam, bam, or cxb files for two. Cufflinks Bioinformatics Tutorial.
From www.researchgate.net
Flowchart of the bioinformatic pipeline. The pipeline performs multiple Cufflinks Bioinformatics Tutorial #how does cufflinks assemble transcripts? The paper includes a diagram (figure 2) describing how the various parts of the cufflinks package (and its companion tool tophat) fit together. This tutorial illustrates how to perform basic differential expression test between 2 groups of samples (for instance: Cuffnorm takes a gtf2/gff3 file of transcripts as input, along with two or more sam,. Cufflinks Bioinformatics Tutorial.
From github.com
cufflinks/tutorial.html at master · coletrapnelllab/cufflinks · GitHub Cufflinks Bioinformatics Tutorial The paper includes a diagram (figure 2) describing how the various parts of the cufflinks package (and its companion tool tophat) fit together. #how does cufflinks calculate transcript abundances? This tutorial illustrates how to perform basic differential expression test between 2 groups of samples (for instance: #how does cufflinks assemble transcripts? Cuffnorm takes a gtf2/gff3 file of transcripts as input,. Cufflinks Bioinformatics Tutorial.
From melbournebioinformatics.github.io
Introduction to bulk RNAseq analysis Bioinformatics Documentation Cufflinks Bioinformatics Tutorial The cufflinks suite includes a. #how does cufflinks calculate transcript abundances? #how does cufflinks assemble transcripts? Cuffnorm takes a gtf2/gff3 file of transcripts as input, along with two or more sam, bam, or cxb files for two or more samples. “treated” vs “control”, “disease” vs “control”, “tissue a” vs “tissue b”. The paper includes a diagram (figure 2) describing how. Cufflinks Bioinformatics Tutorial.
From www.youtube.com
How to create Cufflinks in rhino 3d 023 Rhino Tutorial 023 Cufflink Cufflinks Bioinformatics Tutorial #how does cufflinks assemble transcripts? This tutorial illustrates how to perform basic differential expression test between 2 groups of samples (for instance: Cuffnorm takes a gtf2/gff3 file of transcripts as input, along with two or more sam, bam, or cxb files for two or more samples. “treated” vs “control”, “disease” vs “control”, “tissue a” vs “tissue b”. #how does cufflinks. Cufflinks Bioinformatics Tutorial.
From github.com
GitHub MADscientist314/microbiome_bioinformatics_2020_tutorial_series Cufflinks Bioinformatics Tutorial This tutorial illustrates how to perform basic differential expression test between 2 groups of samples (for instance: “treated” vs “control”, “disease” vs “control”, “tissue a” vs “tissue b”. #how does cufflinks calculate transcript abundances? The paper includes a diagram (figure 2) describing how the various parts of the cufflinks package (and its companion tool tophat) fit together. #how does cufflinks. Cufflinks Bioinformatics Tutorial.
From www.youtube.com
Nextflow for Bioinformatics Tutorial Episode 3 Indexing Genomes Cufflinks Bioinformatics Tutorial The paper includes a diagram (figure 2) describing how the various parts of the cufflinks package (and its companion tool tophat) fit together. #how does cufflinks calculate transcript abundances? “treated” vs “control”, “disease” vs “control”, “tissue a” vs “tissue b”. The cufflinks suite includes a. This tutorial illustrates how to perform basic differential expression test between 2 groups of samples. Cufflinks Bioinformatics Tutorial.
From www.outfittrends.com
How to Put on Cufflinks? 10 Simple Tips with Tutorial Cufflinks Bioinformatics Tutorial This tutorial illustrates how to perform basic differential expression test between 2 groups of samples (for instance: Cuffnorm takes a gtf2/gff3 file of transcripts as input, along with two or more sam, bam, or cxb files for two or more samples. The cufflinks suite includes a. The paper includes a diagram (figure 2) describing how the various parts of the. Cufflinks Bioinformatics Tutorial.
From support.mathworks.com
Cufflinks Support Package for the Bioinformatics Toolbox File Cufflinks Bioinformatics Tutorial The paper includes a diagram (figure 2) describing how the various parts of the cufflinks package (and its companion tool tophat) fit together. The cufflinks suite includes a. This tutorial illustrates how to perform basic differential expression test between 2 groups of samples (for instance: #how does cufflinks assemble transcripts? “treated” vs “control”, “disease” vs “control”, “tissue a” vs “tissue. Cufflinks Bioinformatics Tutorial.
From slideplayer.com
Canadian Bioinformatics ppt download Cufflinks Bioinformatics Tutorial Cuffnorm takes a gtf2/gff3 file of transcripts as input, along with two or more sam, bam, or cxb files for two or more samples. “treated” vs “control”, “disease” vs “control”, “tissue a” vs “tissue b”. #how does cufflinks assemble transcripts? The cufflinks suite includes a. The paper includes a diagram (figure 2) describing how the various parts of the cufflinks. Cufflinks Bioinformatics Tutorial.
From www.youtube.com
What is bioinformatics bioinformatics for beginners bioinformatics Cufflinks Bioinformatics Tutorial The cufflinks suite includes a. Cuffnorm takes a gtf2/gff3 file of transcripts as input, along with two or more sam, bam, or cxb files for two or more samples. This tutorial illustrates how to perform basic differential expression test between 2 groups of samples (for instance: “treated” vs “control”, “disease” vs “control”, “tissue a” vs “tissue b”. The paper includes. Cufflinks Bioinformatics Tutorial.
From training.galaxyproject.org
Handson Referencebased RNAseq data analysis (long) / Transcriptomics Cufflinks Bioinformatics Tutorial “treated” vs “control”, “disease” vs “control”, “tissue a” vs “tissue b”. #how does cufflinks assemble transcripts? This tutorial illustrates how to perform basic differential expression test between 2 groups of samples (for instance: Cuffnorm takes a gtf2/gff3 file of transcripts as input, along with two or more sam, bam, or cxb files for two or more samples. #how does cufflinks. Cufflinks Bioinformatics Tutorial.
From www.youtube.com
Tutorial NeoPixel Novelty Cufflinks with LEDs and Scrolling Text YouTube Cufflinks Bioinformatics Tutorial The paper includes a diagram (figure 2) describing how the various parts of the cufflinks package (and its companion tool tophat) fit together. Cuffnorm takes a gtf2/gff3 file of transcripts as input, along with two or more sam, bam, or cxb files for two or more samples. “treated” vs “control”, “disease” vs “control”, “tissue a” vs “tissue b”. #how does. Cufflinks Bioinformatics Tutorial.
From ww2.mathworks.cn
Build and run bioinformatics pipelines MATLAB MathWorks 中国 Cufflinks Bioinformatics Tutorial #how does cufflinks assemble transcripts? The cufflinks suite includes a. The paper includes a diagram (figure 2) describing how the various parts of the cufflinks package (and its companion tool tophat) fit together. “treated” vs “control”, “disease” vs “control”, “tissue a” vs “tissue b”. #how does cufflinks calculate transcript abundances? Cuffnorm takes a gtf2/gff3 file of transcripts as input, along. Cufflinks Bioinformatics Tutorial.
From www.slideserve.com
PPT Introduction to Bioinformatics Tutorial no. 12 PowerPoint Cufflinks Bioinformatics Tutorial The cufflinks suite includes a. Cuffnorm takes a gtf2/gff3 file of transcripts as input, along with two or more sam, bam, or cxb files for two or more samples. “treated” vs “control”, “disease” vs “control”, “tissue a” vs “tissue b”. #how does cufflinks calculate transcript abundances? This tutorial illustrates how to perform basic differential expression test between 2 groups of. Cufflinks Bioinformatics Tutorial.
From www.researchgate.net
A schematic representation of a workflow for bioinformatics analysis Cufflinks Bioinformatics Tutorial “treated” vs “control”, “disease” vs “control”, “tissue a” vs “tissue b”. The cufflinks suite includes a. This tutorial illustrates how to perform basic differential expression test between 2 groups of samples (for instance: #how does cufflinks assemble transcripts? Cuffnorm takes a gtf2/gff3 file of transcripts as input, along with two or more sam, bam, or cxb files for two or. Cufflinks Bioinformatics Tutorial.
From www.youtube.com
Bringing Bioinformatics Tutorials to Life with sandbox.bio YouTube Cufflinks Bioinformatics Tutorial The paper includes a diagram (figure 2) describing how the various parts of the cufflinks package (and its companion tool tophat) fit together. The cufflinks suite includes a. Cuffnorm takes a gtf2/gff3 file of transcripts as input, along with two or more sam, bam, or cxb files for two or more samples. “treated” vs “control”, “disease” vs “control”, “tissue a”. Cufflinks Bioinformatics Tutorial.
From www.researchgate.net
Bioinformatics steps. (A) Heatmap analysis shows differences in gene Cufflinks Bioinformatics Tutorial The cufflinks suite includes a. The paper includes a diagram (figure 2) describing how the various parts of the cufflinks package (and its companion tool tophat) fit together. #how does cufflinks calculate transcript abundances? “treated” vs “control”, “disease” vs “control”, “tissue a” vs “tissue b”. This tutorial illustrates how to perform basic differential expression test between 2 groups of samples. Cufflinks Bioinformatics Tutorial.
From cole-trapnell-lab.github.io
Cufflinks Cufflinks Bioinformatics Tutorial “treated” vs “control”, “disease” vs “control”, “tissue a” vs “tissue b”. The cufflinks suite includes a. #how does cufflinks assemble transcripts? #how does cufflinks calculate transcript abundances? This tutorial illustrates how to perform basic differential expression test between 2 groups of samples (for instance: Cuffnorm takes a gtf2/gff3 file of transcripts as input, along with two or more sam, bam,. Cufflinks Bioinformatics Tutorial.
From www.bioinformatics.org
Multiple sequence alignment Strap Cufflinks Bioinformatics Tutorial This tutorial illustrates how to perform basic differential expression test between 2 groups of samples (for instance: Cuffnorm takes a gtf2/gff3 file of transcripts as input, along with two or more sam, bam, or cxb files for two or more samples. #how does cufflinks assemble transcripts? The paper includes a diagram (figure 2) describing how the various parts of the. Cufflinks Bioinformatics Tutorial.
From www.researchgate.net
Workflow of the bioinformatics analysis and the results of mapping and Cufflinks Bioinformatics Tutorial Cuffnorm takes a gtf2/gff3 file of transcripts as input, along with two or more sam, bam, or cxb files for two or more samples. #how does cufflinks calculate transcript abundances? The paper includes a diagram (figure 2) describing how the various parts of the cufflinks package (and its companion tool tophat) fit together. This tutorial illustrates how to perform basic. Cufflinks Bioinformatics Tutorial.
From morioh.com
Data Visualization using Plotly & Cufflinks in English Python Tutorial Cufflinks Bioinformatics Tutorial #how does cufflinks assemble transcripts? Cuffnorm takes a gtf2/gff3 file of transcripts as input, along with two or more sam, bam, or cxb files for two or more samples. “treated” vs “control”, “disease” vs “control”, “tissue a” vs “tissue b”. The paper includes a diagram (figure 2) describing how the various parts of the cufflinks package (and its companion tool. Cufflinks Bioinformatics Tutorial.