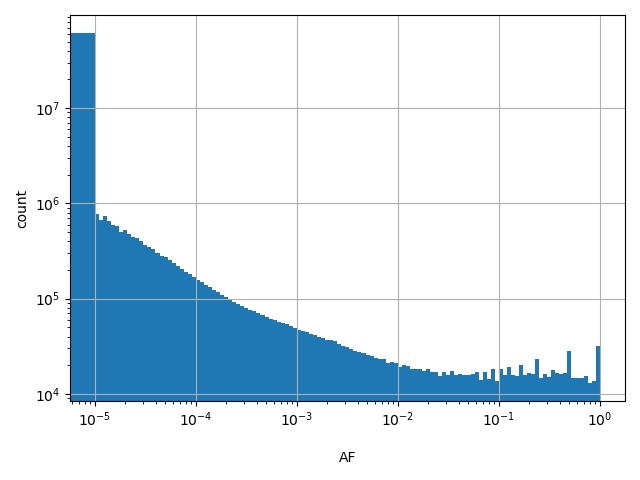
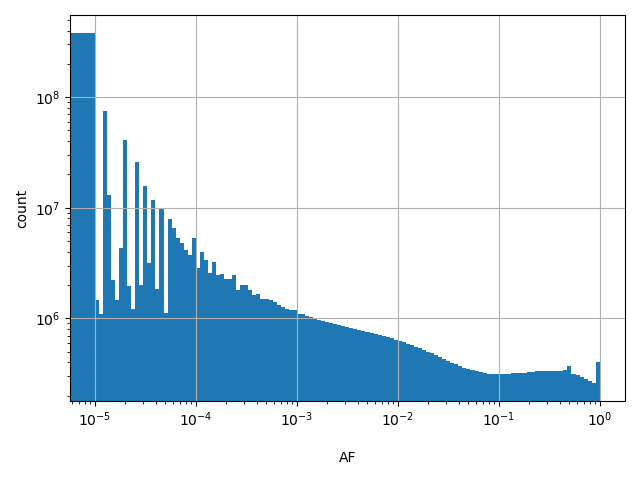
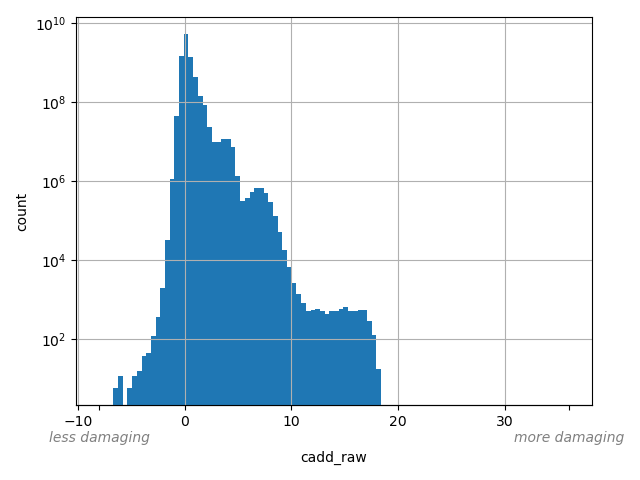
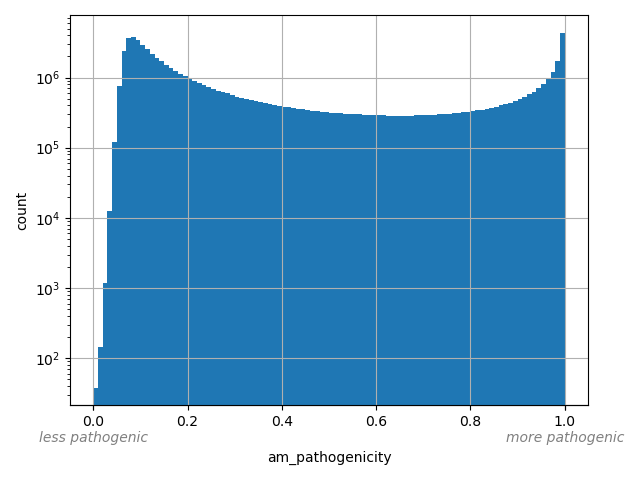
dbSNP ID (i.e. rs number)
position_aggregator: mean [default]
allele_aggregator: max [default]

Annotator to use with scores that depend on allele like variant frequencies, etc.
- input_annotatable:
normalized_allele