Distribution Analysis
Ann Wells
April 23, 2023
Introduction and Data files
R Markdown
This dataset contains nine tissues (heart, hippocampus, hypothalamus, kidney, liver, prefrontal cortex, skeletal muscle, small intestine, and spleen) from C57BL/6J mice that were fed 2-deoxyglucose (6g/L) through their drinking water for 96hrs or 4wks. Organs from mice were harvested and processed for metabolomics and transcriptomics. The data in this document pertains to the transcriptomics data only. This document specifically calculates and plots the distributions for log transformed data. The counts that were used were FPKM normalized before being log transformed.
needed.packages <- c("tidyverse", "here", "functional", "gplots", "dplyr", "GeneOverlap", "R.utils", "reshape2","magrittr","data.table", "RColorBrewer","preprocessCore")
for(i in 1:length(needed.packages)){library(needed.packages[i], character.only = TRUE)}Read in data
tdata.FPKM.sample.info <- readRDS(here("Data","20190406_RNAseq_B6_4wk_2DG_counts_phenotypes.RData"))
tdata.FPKM <- readRDS(here("Data","20190406_RNAseq_B6_4wk_2DG_counts_numeric.RData"))Boxplots
These plots will show the distribution of the data by displaying the variability or dispersion of the data.
Time
This boxplot groups control and treated samples for each time point and tissue.
tdata.FPKM.sample.info <- readRDS(here("Data","20190406_RNAseq_B6_4wk_2DG_counts_phenotypes.RData"))
tdata.FPKM <- readRDS(here("Data","20190406_RNAseq_B6_4wk_2DG_counts_numeric.RData"))
log.tdata.FPKM <- log(tdata.FPKM + 1)
log.tdata.FPKM.sample.info <- cbind(log.tdata.FPKM, tdata.FPKM.sample.info[,27238:27240])
log.tdata.FPKM.sample.info <- rownames_to_column(as.data.frame(log.tdata.FPKM.sample.info))
melttissue <- melt(log.tdata.FPKM.sample.info, variable_name="Tissue")
ggplot(melttissue, aes(x = Tissue, y = value, fill = Time)) + geom_boxplot() + xlab("") + ylab(expression(log(count + 1))) + scale_fill_manual(values = c("darkred", "dodgerblue")) + theme(axis.text.x = element_text(angle = 45))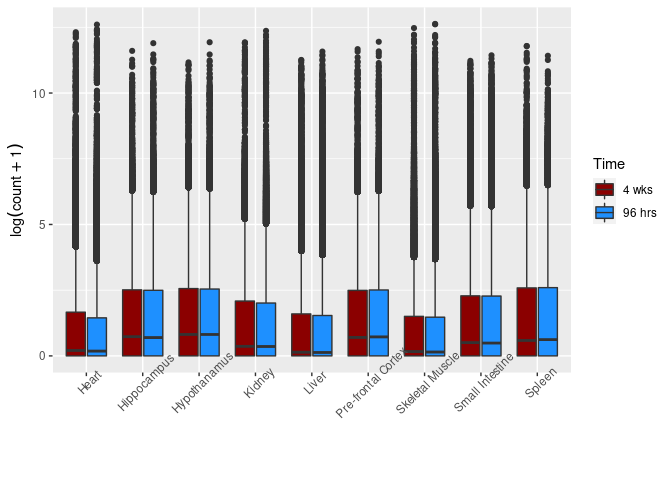
Tissue
This boxplot groups control and treated samples for each time point and tissue but displays the data by time point.
tdata.FPKM.sample.info <- readRDS(here("Data","20190406_RNAseq_B6_4wk_2DG_counts_phenotypes.RData"))
tdata.FPKM <- readRDS(here("Data","20190406_RNAseq_B6_4wk_2DG_counts_numeric.RData"))
log.tdata.FPKM <- log(tdata.FPKM + 1)
log.tdata.FPKM.sample.info <- cbind(log.tdata.FPKM, tdata.FPKM.sample.info[,27238:27240])
log.tdata.FPKM.sample.info <- rownames_to_column(as.data.frame(log.tdata.FPKM.sample.info))
melttissue <- melt(log.tdata.FPKM.sample.info, variable_name="Tissue")
ggplot(melttissue, aes(x = Time, y = value, fill = Tissue)) + geom_boxplot() + xlab("") + ylab(expression(log(count + 1))) + theme(axis.text.x = element_text(angle = 45))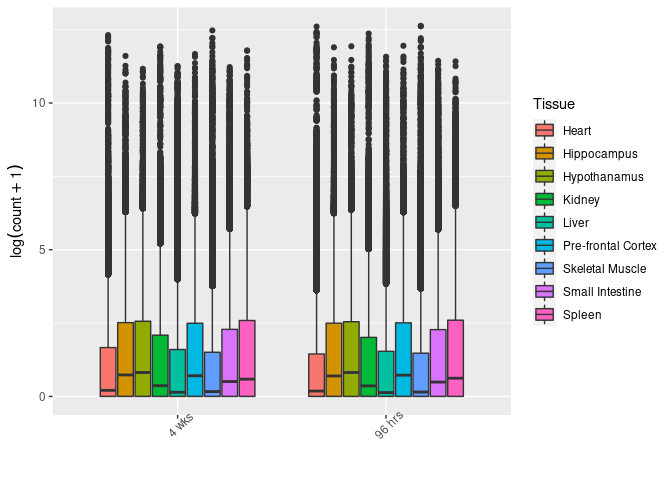
Treatment
This boxplot groups time points for each treatment and tissue.
tdata.FPKM.sample.info <- readRDS(here("Data","20190406_RNAseq_B6_4wk_2DG_counts_phenotypes.RData"))
tdata.FPKM <- readRDS(here("Data","20190406_RNAseq_B6_4wk_2DG_counts_numeric.RData"))
log.tdata.FPKM <- log(tdata.FPKM + 1)
log.tdata.FPKM.sample.info <- cbind(log.tdata.FPKM, tdata.FPKM.sample.info[,27238:27240])
log.tdata.FPKM.sample.info <- rownames_to_column(as.data.frame(log.tdata.FPKM.sample.info))
melttissue <- melt(log.tdata.FPKM.sample.info, variable_name="Tissue")
ggplot(melttissue, aes(x = Tissue, y = value, fill = Treatment)) + geom_boxplot() + xlab("") + ylab(expression(log(count + 1))) + scale_fill_manual(values = c("darkred", "dodgerblue")) + theme(axis.text.x = element_text(angle = 45))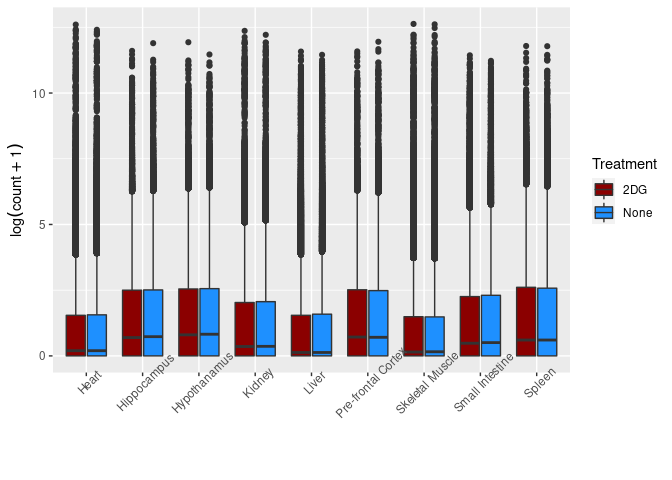
Treatment by tissue by time
This boxplot displays treatment and time for each tissue.
tdata.FPKM.sample.info <- readRDS(here("Data","20190406_RNAseq_B6_4wk_2DG_counts_phenotypes.RData"))
tdata.FPKM <- readRDS(here("Data","20190406_RNAseq_B6_4wk_2DG_counts_numeric.RData"))
log.tdata.FPKM <- log(tdata.FPKM + 1)
log.tdata.FPKM.sample.info <- cbind(log.tdata.FPKM, tdata.FPKM.sample.info[,27238:27240])
log.tdata.FPKM.sample.info <- rownames_to_column(as.data.frame(log.tdata.FPKM.sample.info))
melttissue <- melt(log.tdata.FPKM.sample.info, variable_name="Tissue")
ggplot(melttissue, aes(x = Tissue, y = value, fill = Tissue)) + geom_boxplot() + xlab("") + ylab(expression(log(count + 1))) + theme(axis.text.x = element_text(angle = 45)) + facet_wrap(~ Time*Treatment) + theme_dark() + theme(strip.background = element_rect(color="black", size=1.5, linetype="solid")) + theme(axis.text.x = element_text(angle = 90, face = "bold", size = 8,hjust=0.95,vjust=0.2))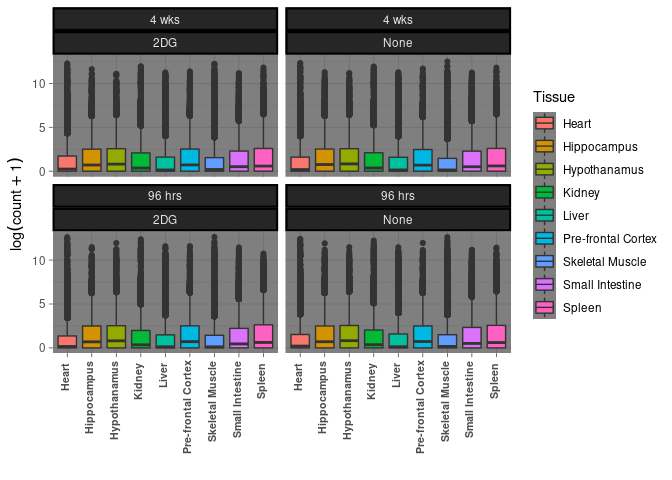
Samples
This boxplot displays all of the samples but they are colored by tissue.
tdata.FPKM.sample.info <- readRDS(here("Data","20190406_RNAseq_B6_4wk_2DG_counts_phenotypes.RData"))
tdata.FPKM <- readRDS(here("Data","20190406_RNAseq_B6_4wk_2DG_counts_numeric.RData"))
log.tdata.FPKM <- log(tdata.FPKM + 1)
log.tdata.FPKM.sample.info <- cbind(log.tdata.FPKM, tdata.FPKM.sample.info[,27238:27240])
log.tdata.FPKM.sample.info <- rownames_to_column(as.data.frame(log.tdata.FPKM.sample.info))
melttissue <- melt(log.tdata.FPKM.sample.info, variable_name="Tissue")
ggplot(melttissue, aes(x = rowname, y = value, fill = Tissue)) + geom_boxplot() + xlab("") + ylab(expression(log(count + 1))) + theme(legend.position='none') + theme(axis.text.x = element_text(angle = 90, face = "bold", size = 4))
Density Plots
Density plots show us the distribution of the data.
Time by treatment
This plot displays treatment and time for each tissue for each sample.
tdata.FPKM.sample.info <- readRDS(here("Data","20190406_RNAseq_B6_4wk_2DG_counts_phenotypes.RData"))
tdata.FPKM <- readRDS(here("Data","20190406_RNAseq_B6_4wk_2DG_counts_numeric.RData"))
log.tdata.FPKM <- log(tdata.FPKM + 1)
log.tdata.FPKM.sample.info <- cbind(log.tdata.FPKM, tdata.FPKM.sample.info[,27238:27240])
log.tdata.FPKM.sample.info <- rownames_to_column(as.data.frame(log.tdata.FPKM.sample.info))
melttissue <- melt(log.tdata.FPKM.sample.info, variable_name="Tissue")
ggplot(melttissue, aes(x = value, colour = rowname, fill = rowname)) + ylim(c(0, 0.5)) +
geom_density(alpha = 0.05, size = .5) + facet_wrap(~ Time*Treatment) + xlab(expression(log(count + 1))) + theme_dark() + theme(strip.background = element_rect(color="black", size=1.5, linetype="solid")) + theme(legend.position = "none")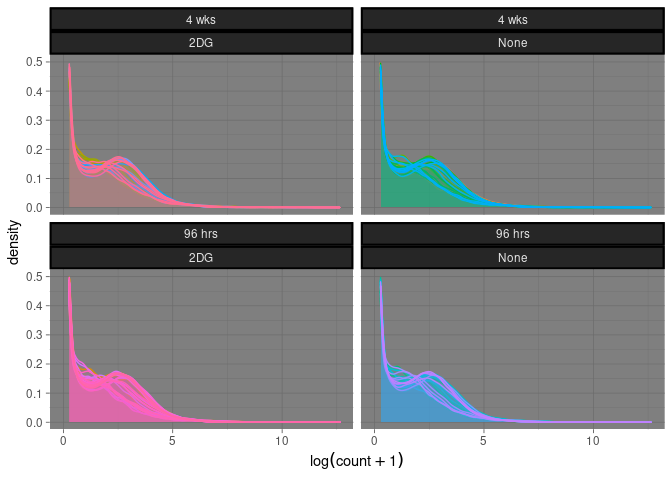
Tissue
This plot groups treatment and time point for each tissue for each sample.
tdata.FPKM.sample.info <- readRDS(here("Data","20190406_RNAseq_B6_4wk_2DG_counts_phenotypes.RData"))
tdata.FPKM <- readRDS(here("Data","20190406_RNAseq_B6_4wk_2DG_counts_numeric.RData"))
log.tdata.FPKM <- log(tdata.FPKM + 1)
log.tdata.FPKM.sample.info <- cbind(log.tdata.FPKM, tdata.FPKM.sample.info[,27238:27240])
log.tdata.FPKM.sample.info <- rownames_to_column(as.data.frame(log.tdata.FPKM.sample.info))
melttissue <- melt(log.tdata.FPKM.sample.info, variable_name="Tissue")
ggplot(melttissue, aes(x = value, colour = Treatment, fill = Time)) + ylim(c(0, 0.5)) +
geom_density(alpha = 0.05, size = .5) + facet_wrap(~ Tissue) + xlab(expression(log(count + 1))) + theme_dark() + theme(strip.background = element_rect(color="black", size=1.5, linetype="solid")) + theme(legend.position = "none")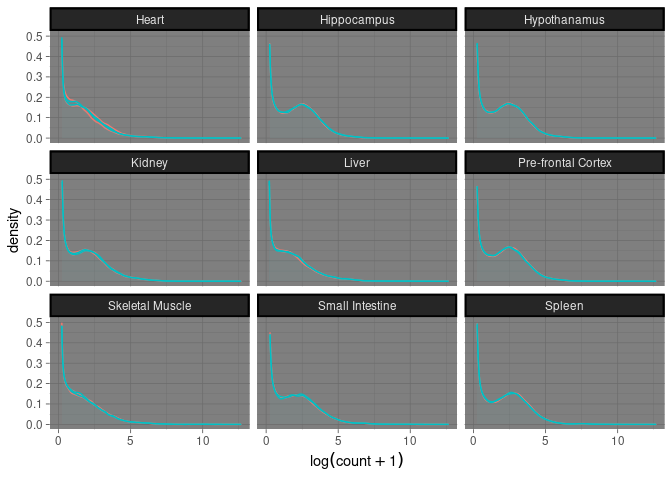
Clustering
This heatmap clusters the data for each sample to determine, which samples are most closely related through their expression.
tdata.FPKM.sample.info <- readRDS(here("Data","20190406_RNAseq_B6_4wk_2DG_counts_phenotypes.RData"))
tdata.FPKM <- readRDS(here("Data","20190406_RNAseq_B6_4wk_2DG_counts_numeric.RData"))
log.tdata.FPKM <- log(tdata.FPKM + 1)
log.tdata.FPKM.sample.info <- cbind(log.tdata.FPKM, tdata.FPKM.sample.info[,27238:27240])
log.data.FPKM.sample.info <- t(sapply(log.tdata.FPKM.sample.info, as.numeric))
colnames(log.data.FPKM.sample.info) <- rownames(log.tdata.FPKM.sample.info)
mat.dist = log.data.FPKM.sample.info
colnames(mat.dist) = paste(colnames(mat.dist), tdata.FPKM.sample.info$Time, tdata.FPKM.sample.info$Treatment, tdata.FPKM.sample.info$Tissue, sep = " : ")
mat.dist = as.matrix(dist(t(mat.dist)))
mat.dist = mat.dist/max(mat.dist)
hmcol = colorRampPalette(brewer.pal(9, "GnBu"))(144)
pheatmap::pheatmap(mat.dist, color = rev(hmcol), fontsize = 2)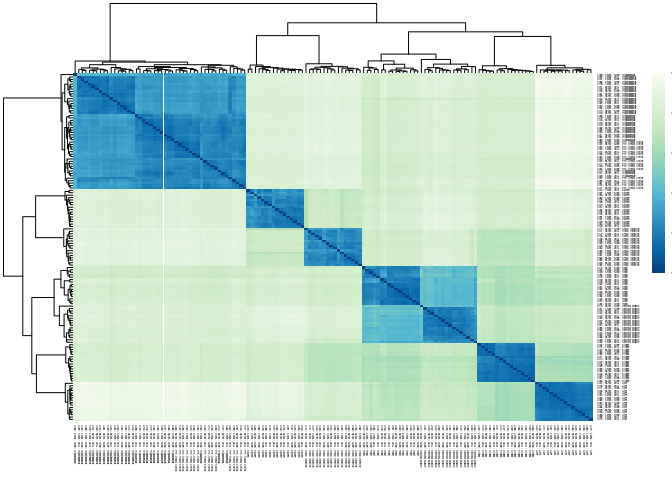
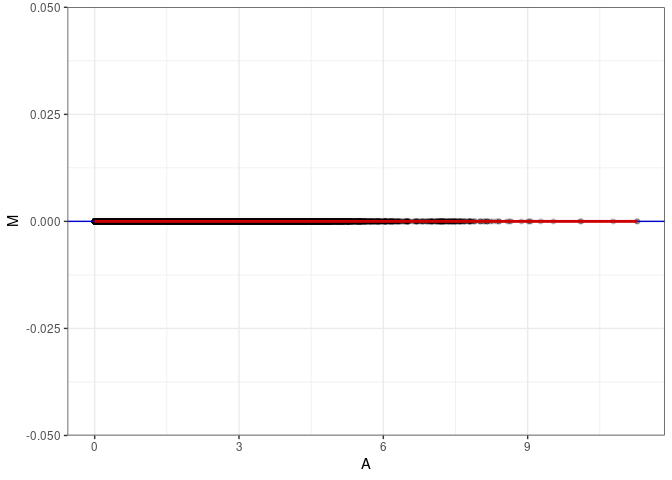
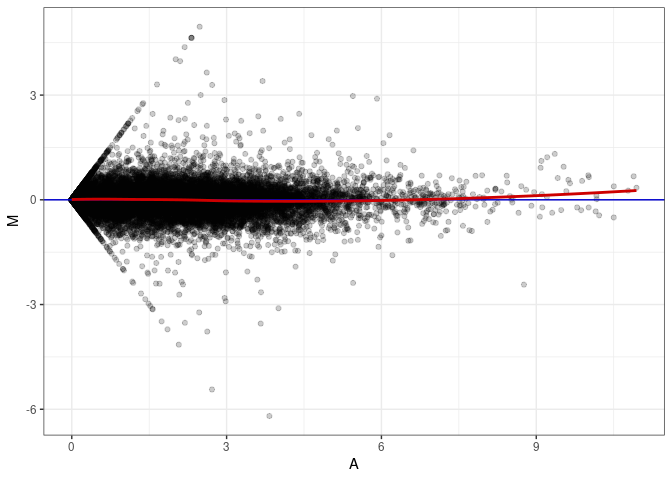
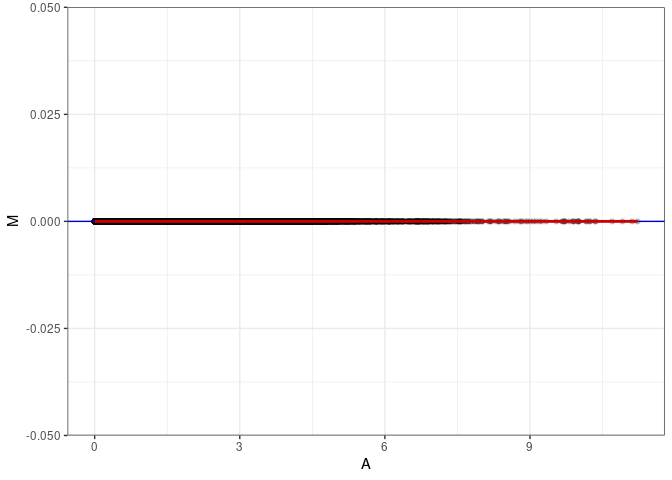
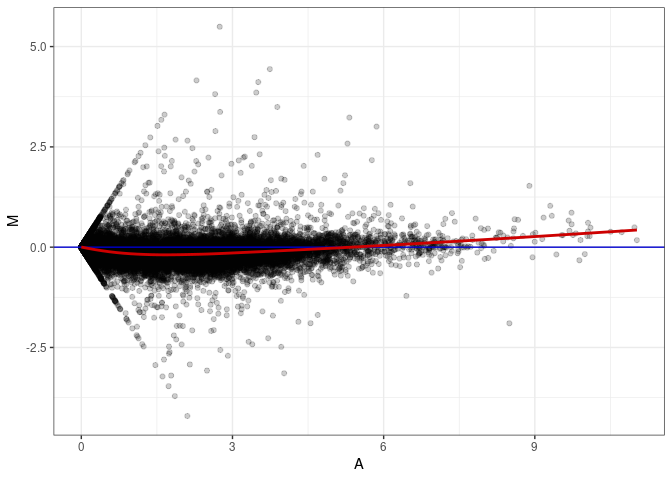
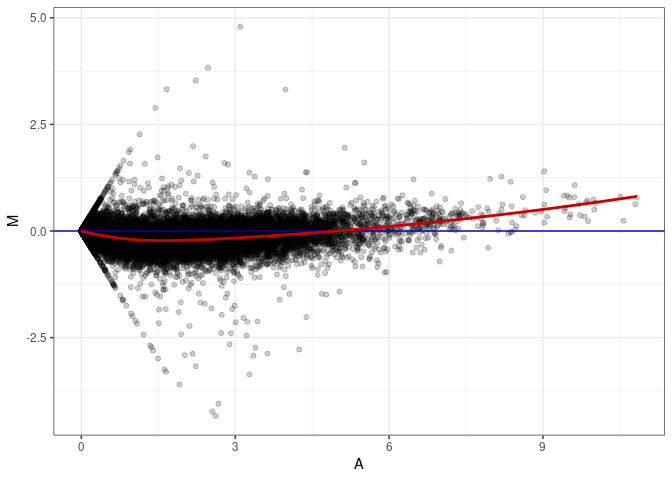
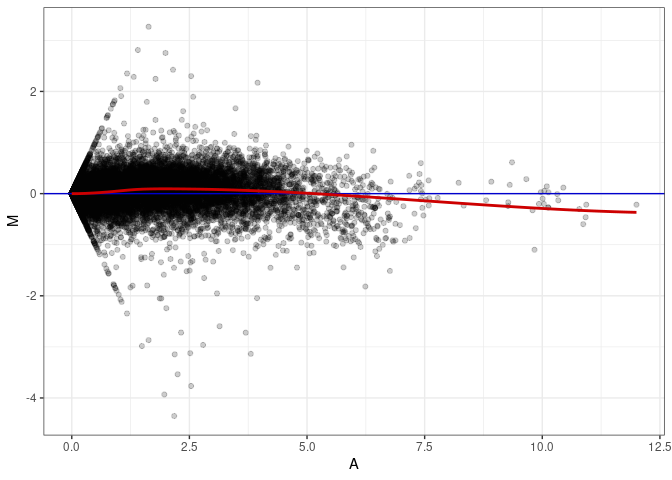
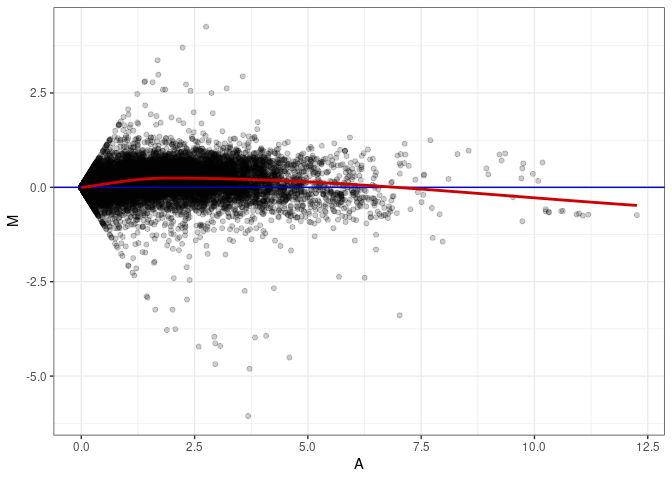
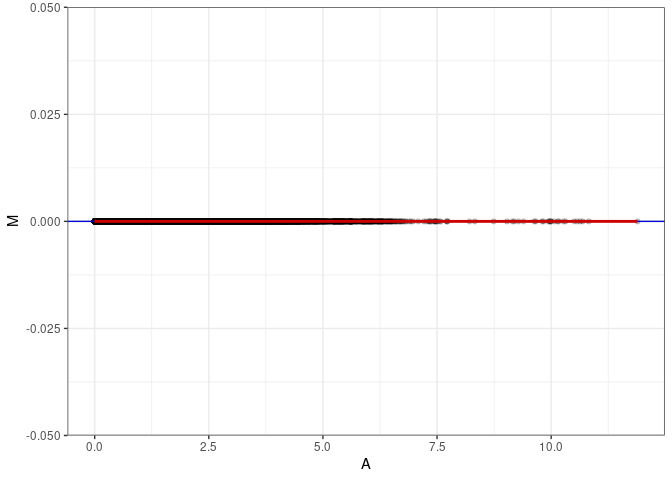
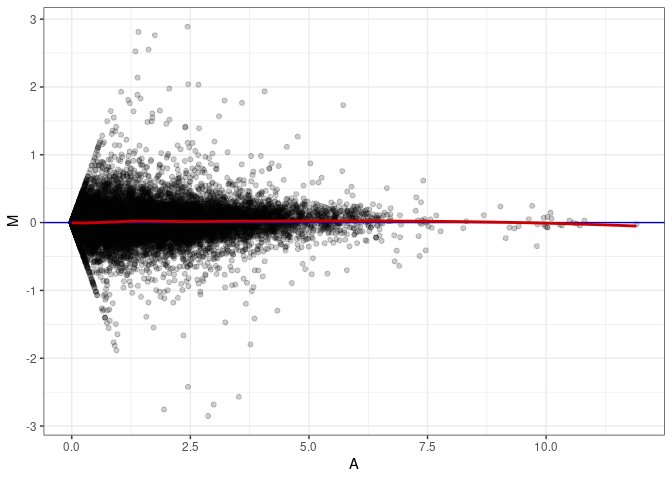
MA plots
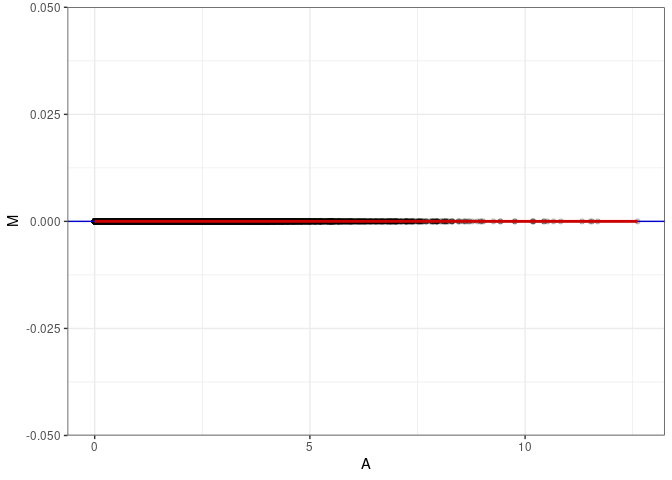
MA plots plot the log-fold change against the log average. The M in the plot is the log of the ratio of counts for each gene between two samples. The A in the plot is the average count for each gene across two samples. Each comparison below is only plotting like with like (i.e. spleen 2DG 4wks with spleen 2DG 4wks). These plots will should hover around zero to visualize that samples within the same treatment and time combination have the same expression profile, relatively.
MA plot for Spleen 2DG 4wks
subdata <- log.tdata.FPKM.sample.info %>% rownames_to_column() %>% filter(Tissue =="Spleen" & Treatment == "2DG" & Time == "4 wks") %>% column_to_rownames()
subdata <- subdata[,-c(27237:27239)]
log.data.FPKM.sample.info <- t(subdata)
p <- c()
for(i in 1:ncol(log.data.FPKM.sample.info)){
for(j in 1:ncol(log.data.FPKM.sample.info)){
x = log.data.FPKM.sample.info[,i]
y = log.data.FPKM.sample.info[,j]
##M-values
M=x-y ##A-values
A = (x + y)/2
df = data.frame(A, M)
cat('###',"MA plot", colnames(log.data.FPKM.sample.info)[[i]], "-", colnames(log.data.FPKM.sample.info)[[j]] ,' \n')
p <- ggplot(df, aes(x = A, y = M)) + geom_point(size = 1.5, alpha = 1/5) +
geom_hline(color = "blue3", yintercept = 0) + stat_smooth(se = FALSE, method = "loess", color = "red3") + theme_bw()
print(p)
#print(htmltools::tagList(p))
cat("\n \n")
}
}MA plot A019 - A019
MA plot A019 - A028
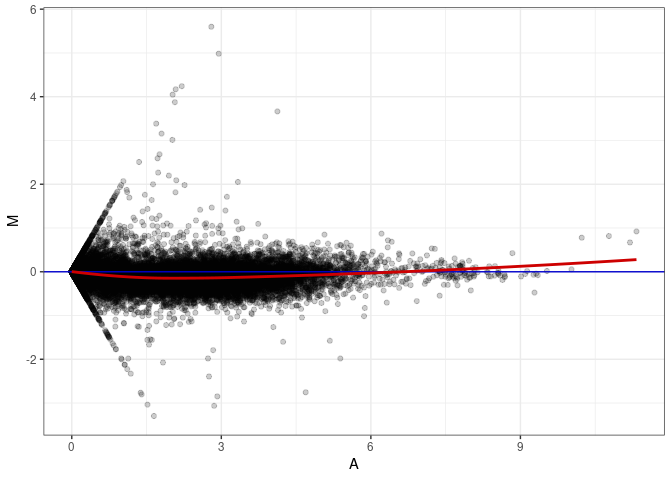
MA plot A019 - A091
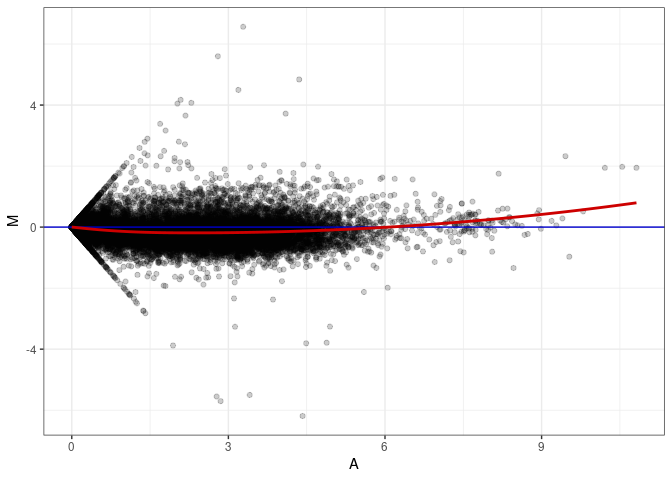
MA plot A019 - A172
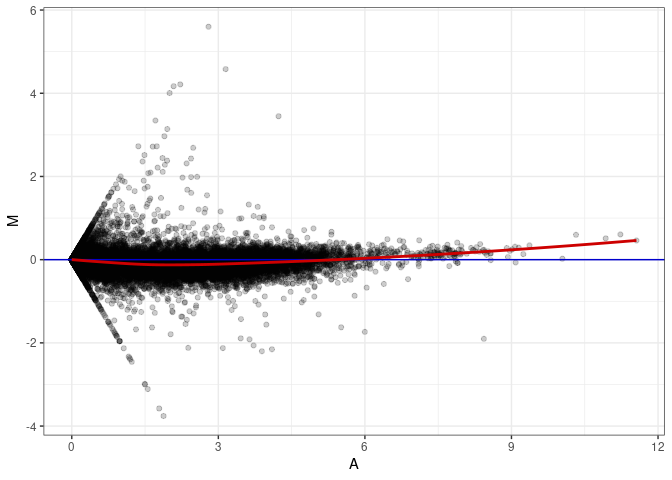
MA plot A028 - A019
MA plot A028 - A028
MA plot A028 - A091
MA plot A028 - A172
MA plot A091 - A019
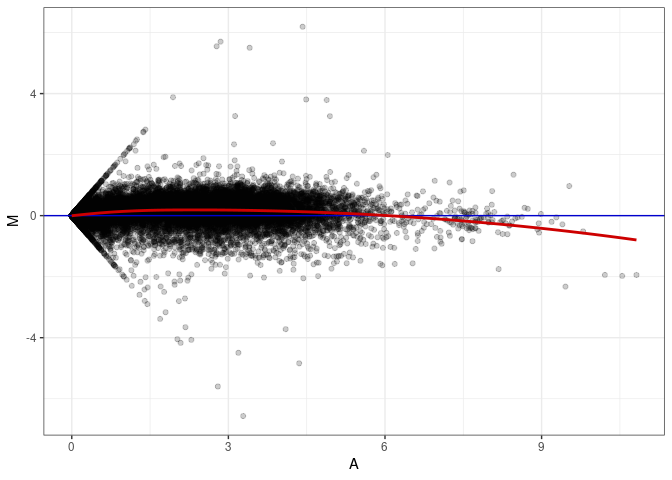
MA plot A091 - A028
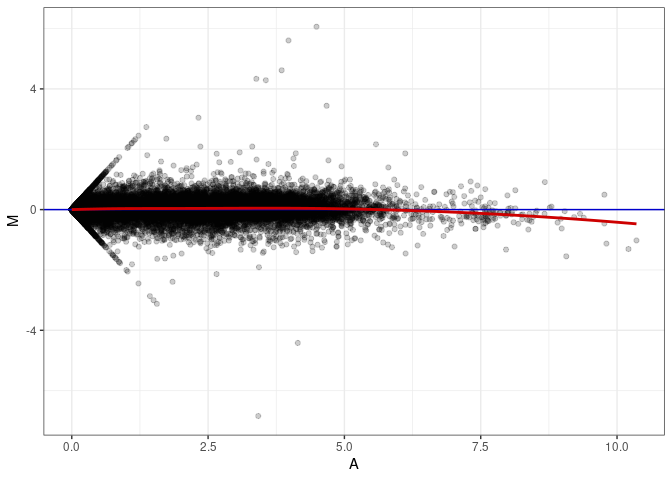
MA plot A091 - A091
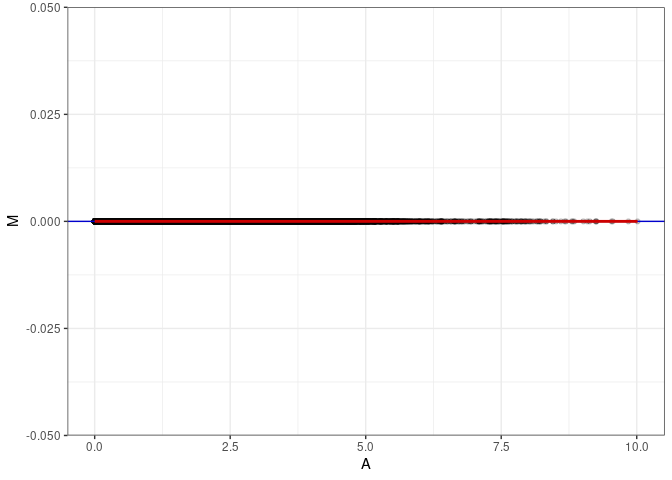
MA plot A091 - A172
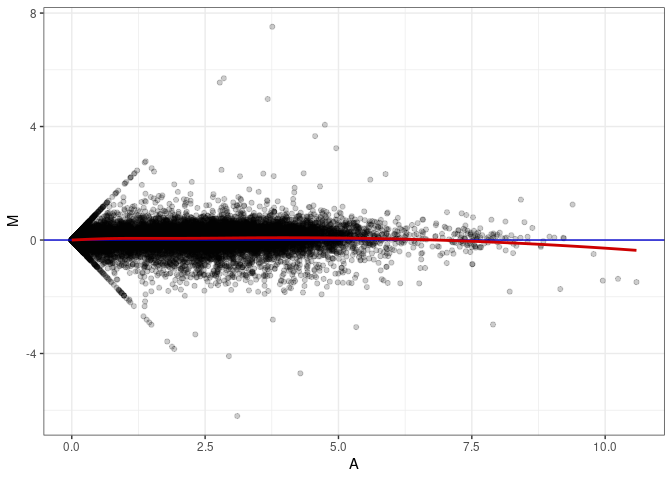
MA plot A172 - A019
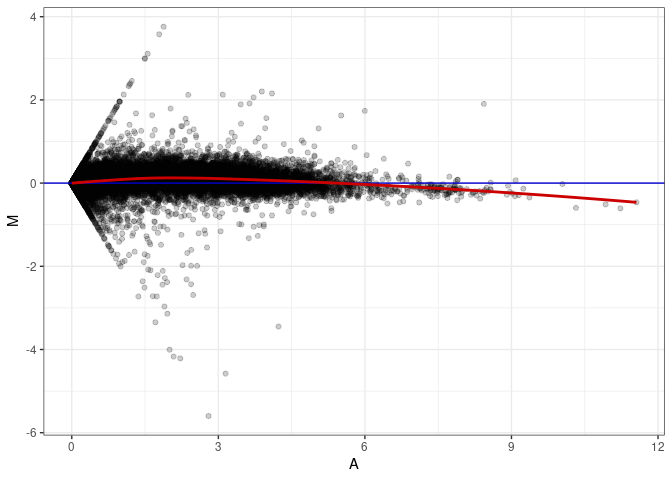
MA plot A172 - A028
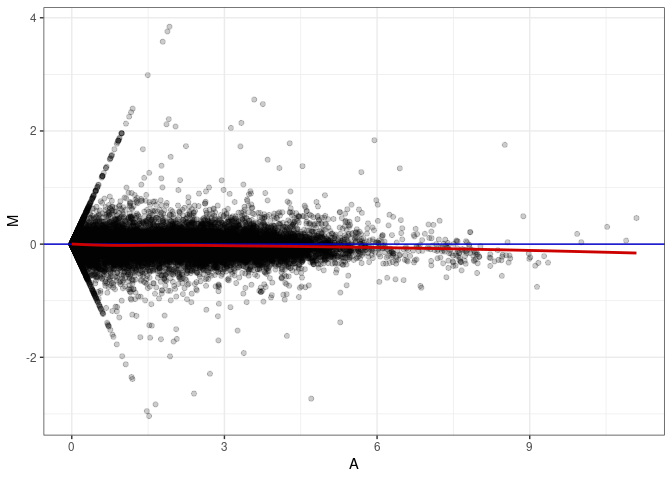
MA plot A172 - A091
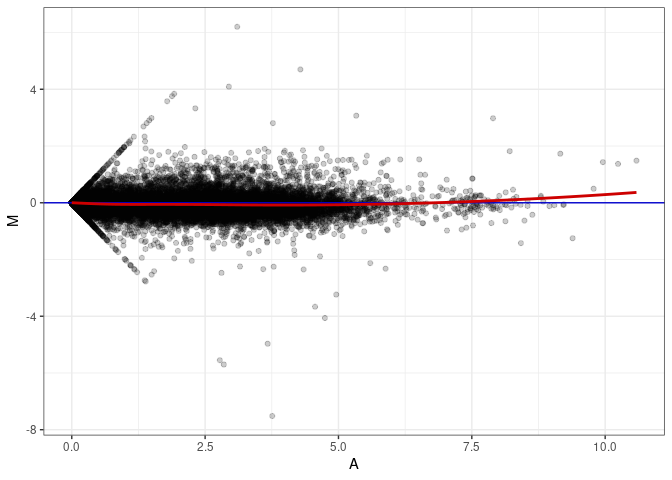
MA plot A172 - A172
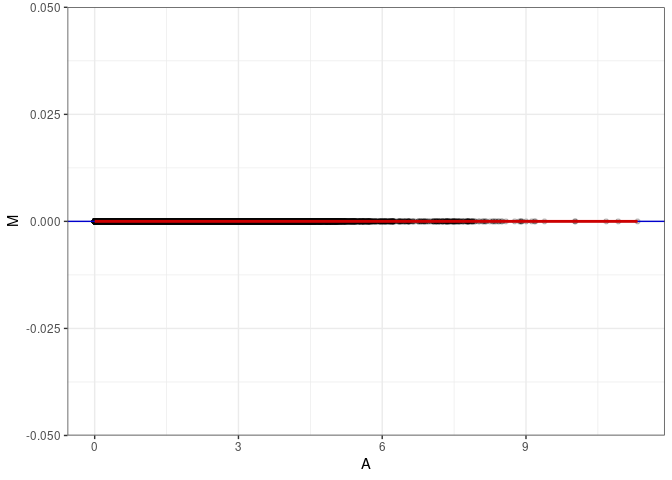
MA plot for Spleen control 4wks
subdata <- log.tdata.FPKM.sample.info %>% rownames_to_column() %>% filter(Tissue =="Spleen" & Treatment == "None" & Time == "4 wks") %>% column_to_rownames()
subdata <- subdata[,-c(27237:27239)]
log.data.FPKM.sample.info <- t(subdata)
for(i in 1:ncol(log.data.FPKM.sample.info)){
for(j in 1:ncol(log.data.FPKM.sample.info)){
x = log.data.FPKM.sample.info[,i]
y = log.data.FPKM.sample.info[,j]
## M-values
M=x-y ## A-values
A = (x + y)/2
df = data.frame(A, M)
cat('###',"MA plot", colnames(log.data.FPKM.sample.info)[[i]], "-", colnames(log.data.FPKM.sample.info)[[j]] ,' \n')
p <- ggplot(df, aes(x = A, y = M)) + geom_point(size = 1.5, alpha = 1/5) +
geom_hline(color = "blue3", yintercept = 0) + stat_smooth(se = FALSE, method = "loess", color = "red3") + theme_bw()
print(p)
#print(htmltools::tagList(ggplotly(p)))
cat(' \n\n')
}
}MA plot A001 - A001
MA plot A001 - A037
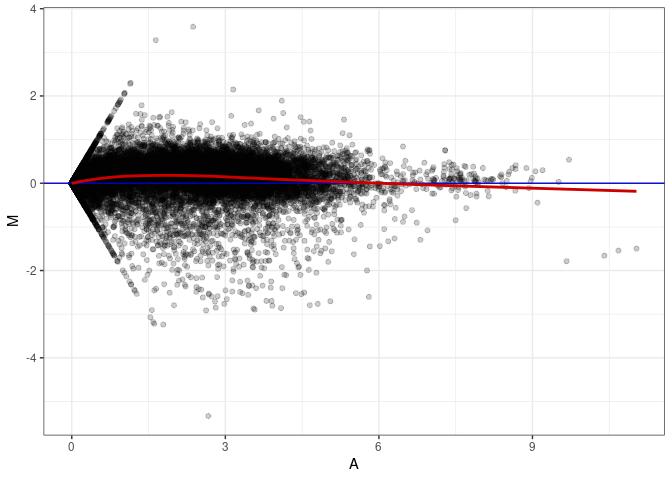
MA plot A001 - A046
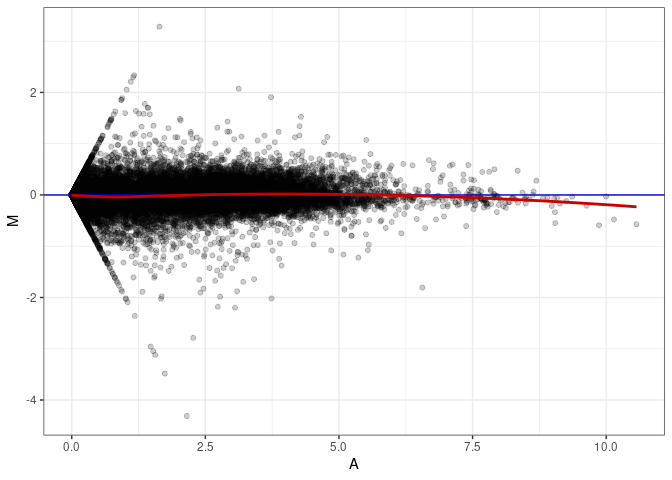
MA plot A001 - A082
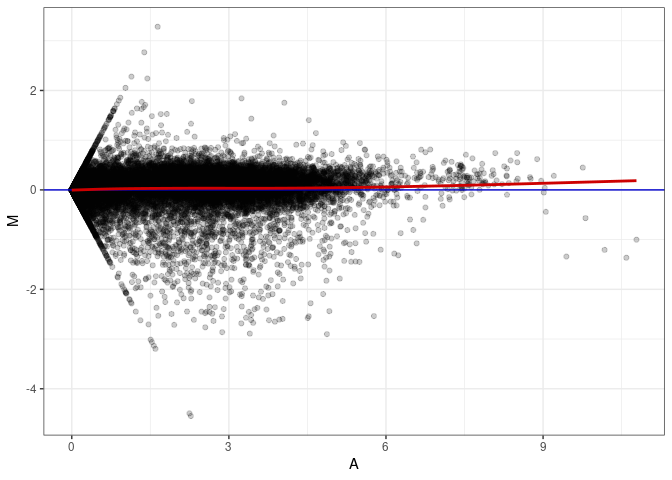
MA plot A037 - A001
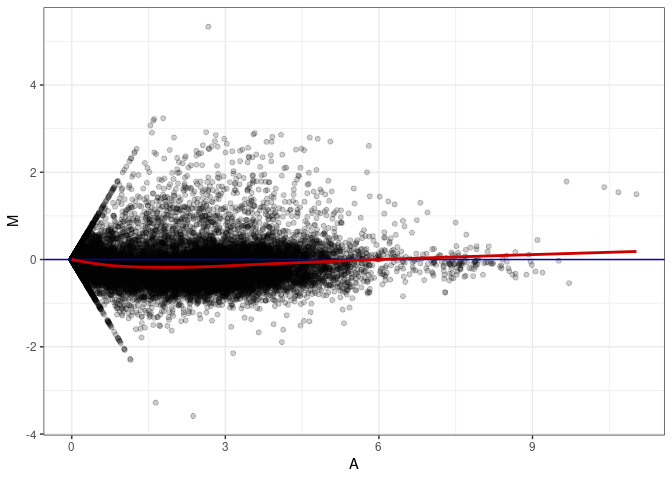
MA plot A037 - A037
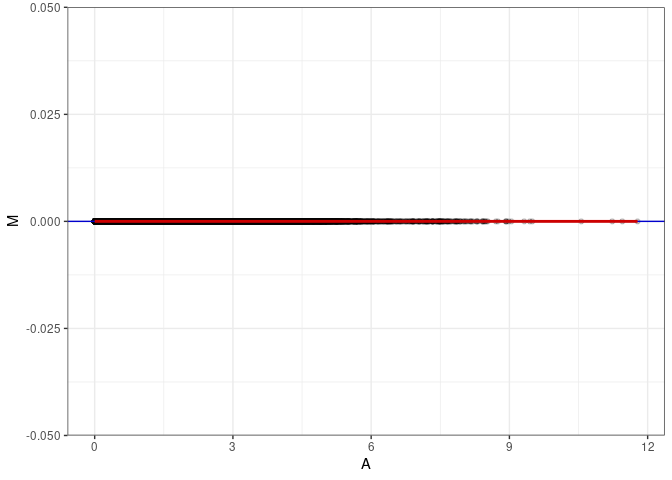
MA plot A037 - A046
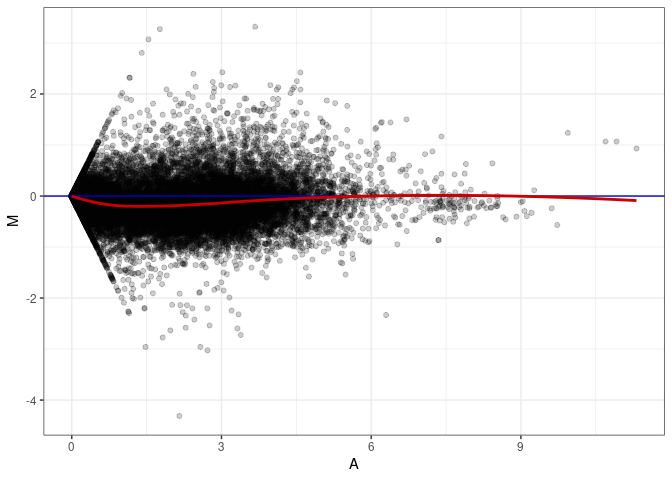
MA plot A037 - A082
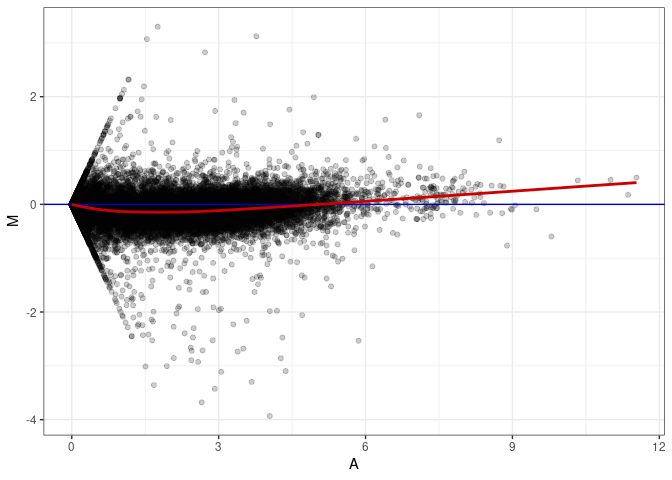
MA plot A046 - A001
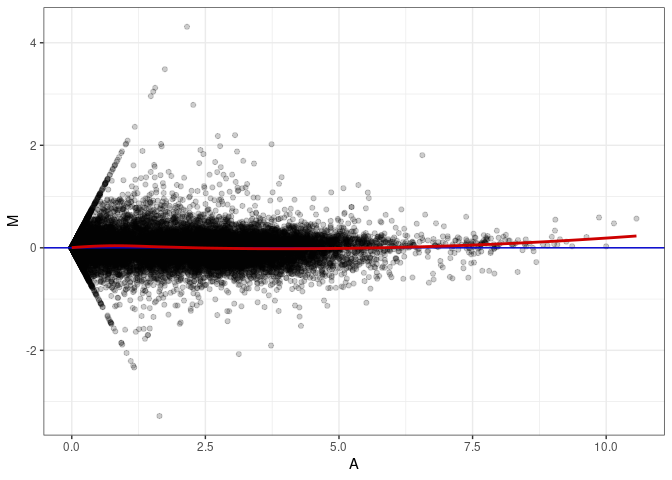
MA plot A046 - A037
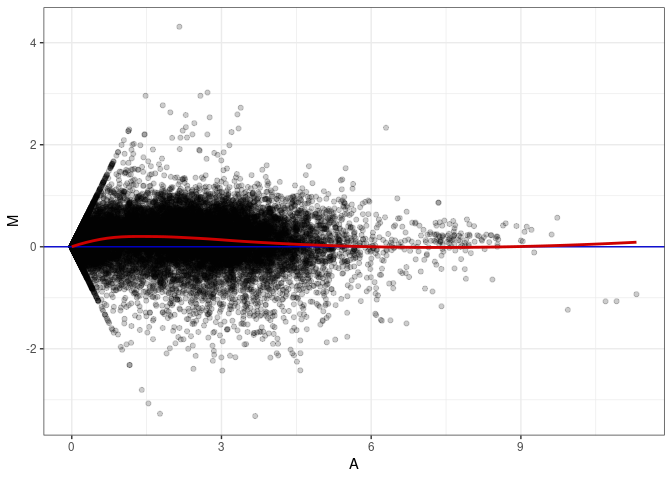
MA plot A046 - A046
MA plot A046 - A082
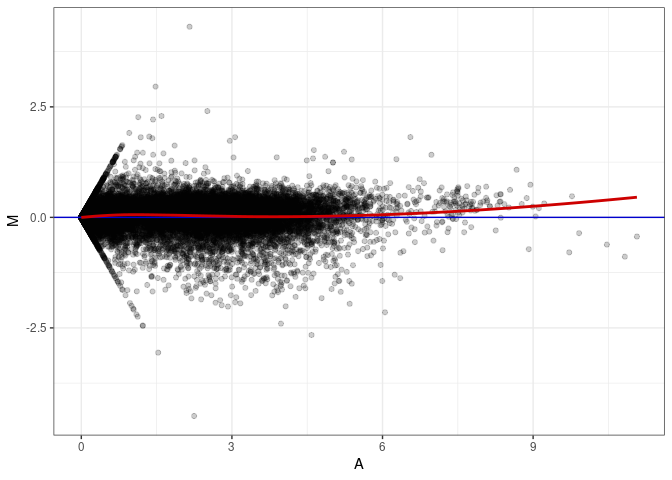
MA plot A082 - A001
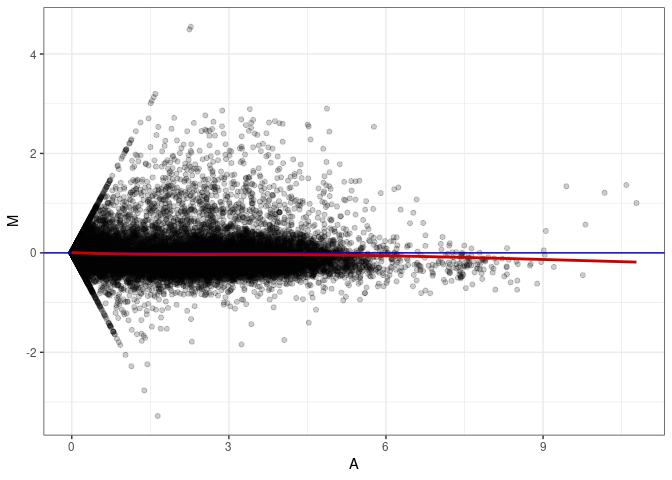
MA plot A082 - A037
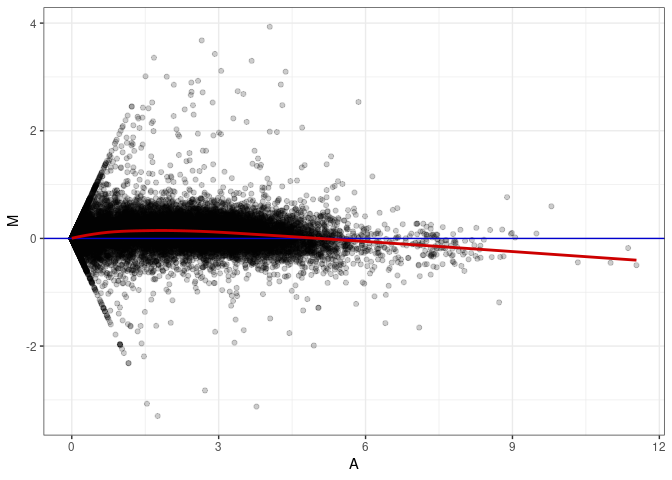
MA plot A082 - A046
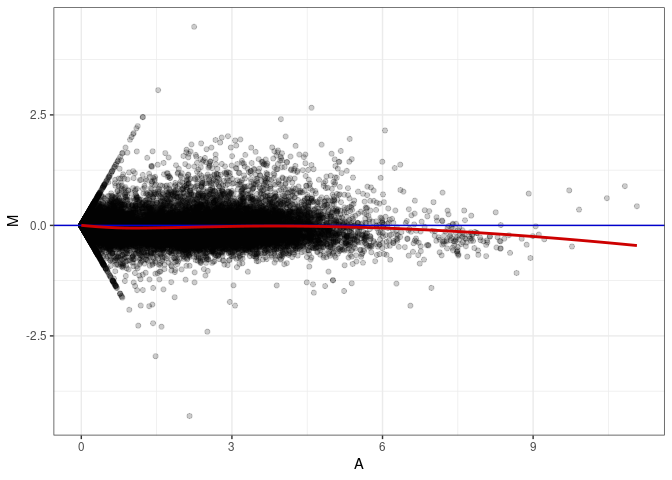
MA plot A082 - A082
MA plot for Spleen 2DG 96hrs
subdata <- log.tdata.FPKM.sample.info %>% rownames_to_column() %>% filter(Tissue =="Spleen" & Treatment == "2DG" & Time == "96 hrs") %>% column_to_rownames()
subdata <- subdata[,-c(27237:27239)]
log.data.FPKM.sample.info <- t(subdata)
for(i in 1:ncol(log.data.FPKM.sample.info)){
for(j in 1:ncol(log.data.FPKM.sample.info)){
x = log.data.FPKM.sample.info[,i]
y = log.data.FPKM.sample.info[,j]
## M-values
M=x-y ## A-values
A = (x + y)/2
df = data.frame(A, M)
cat('###',"MA plot", colnames(log.data.FPKM.sample.info)[[i]], "-", colnames(log.data.FPKM.sample.info)[[j]] ,' \n')
p <- ggplot(df, aes(x = A, y = M)) + geom_point(size = 1.5, alpha = 1/5) +
geom_hline(color = "blue3", yintercept = 0) + stat_smooth(se = FALSE, method = "loess", color = "red3") + theme_bw()
print(p)
#print(htmltools::tagList(ggplotly(p)))
cat("\n \n")
}
}MA plot A010 - A010
MA plot A010 - A136
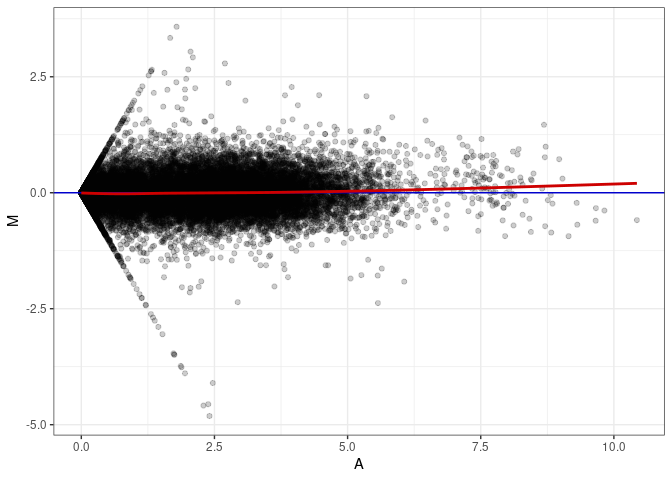
MA plot A010 - A145
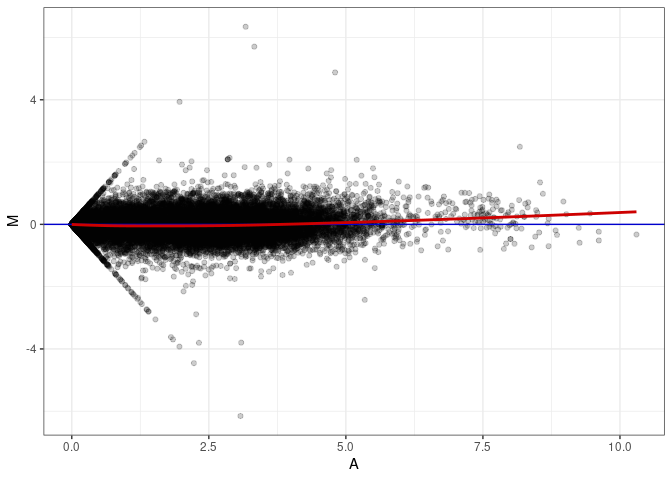
MA plot A010 - A163
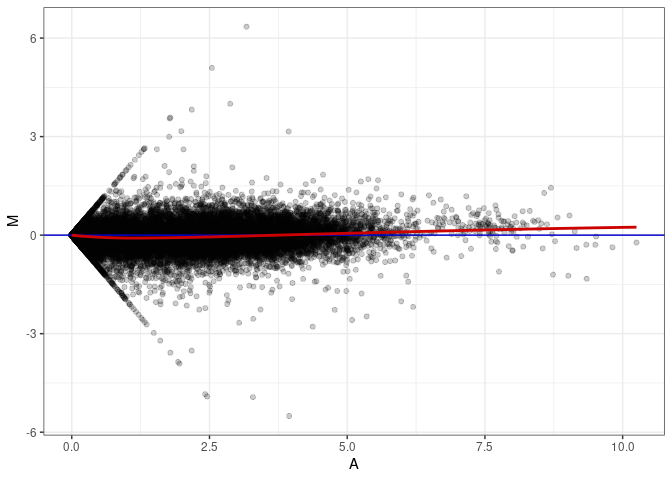
MA plot A136 - A010
MA plot A136 - A136
MA plot A136 - A145
MA plot A136 - A163
MA plot A145 - A010
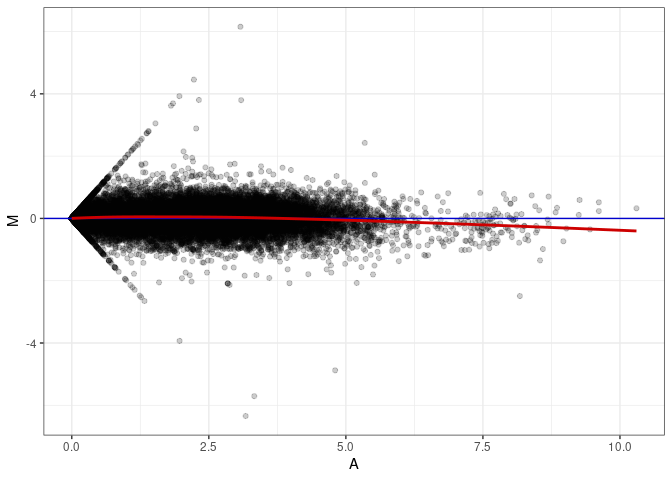
MA plot A145 - A136
MA plot A145 - A145
MA plot A145 - A163
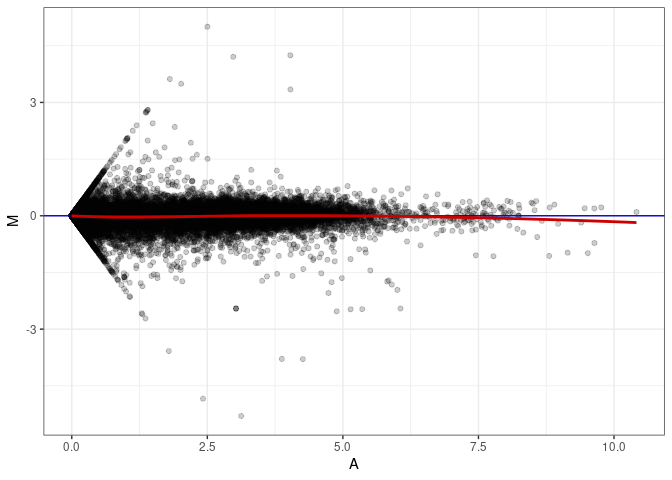
MA plot A163 - A010
MA plot A163 - A136
MA plot A163 - A145
MA plot A163 - A163
MA plot for Spleen control 96hrs
subdata <- log.tdata.FPKM.sample.info %>% rownames_to_column() %>% filter(Tissue =="Spleen" & Treatment == "None" & Time == "96 hrs") %>% column_to_rownames()
subdata <- subdata[,-c(27237:27239)]
log.data.FPKM.sample.info <- t(subdata)
for(i in 1:ncol(log.data.FPKM.sample.info)){
for(j in 1:ncol(log.data.FPKM.sample.info)){
x = log.data.FPKM.sample.info[,i]
y = log.data.FPKM.sample.info[,j]
## M-values
M=x-y ## A-values
A = (x + y)/2
df = data.frame(A, M)
cat('###',"MA plot", colnames(log.data.FPKM.sample.info)[[i]], "-", colnames(log.data.FPKM.sample.info)[[j]] ,' \n')
p <- ggplot(df, aes(x = A, y = M)) + geom_point(size = 1.5, alpha = 1/5) +
geom_hline(color = "blue3", yintercept = 0) + stat_smooth(se = FALSE, method = "loess", color = "red3") + theme_bw()
print(p)
#print(htmltools::tagList(ggplotly(p)))
cat("\n \n")
}
}MA plot A055 - A055
MA plot A055 - A064
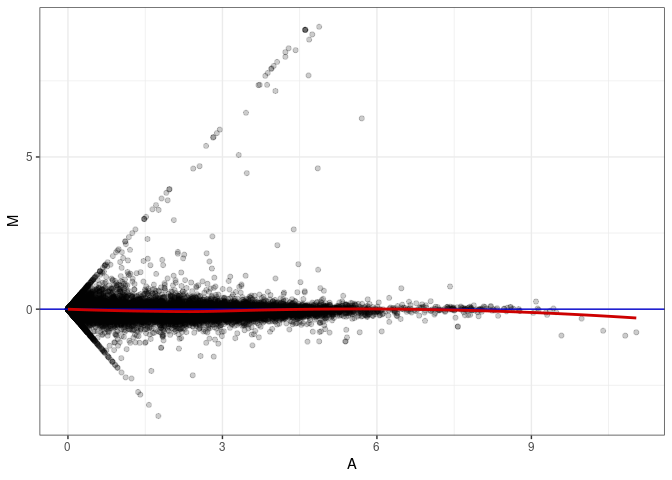
MA plot A055 - A073
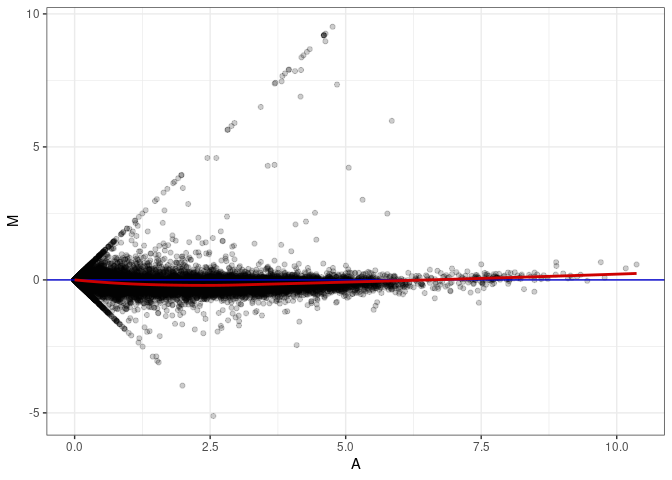
MA plot A055 - A109
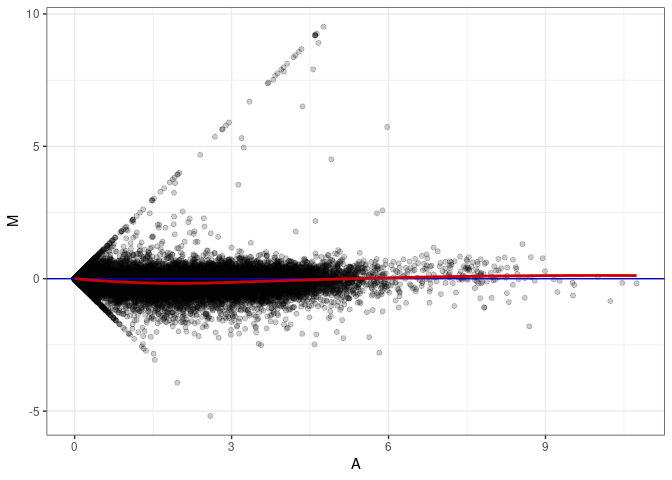
MA plot A064 - A055
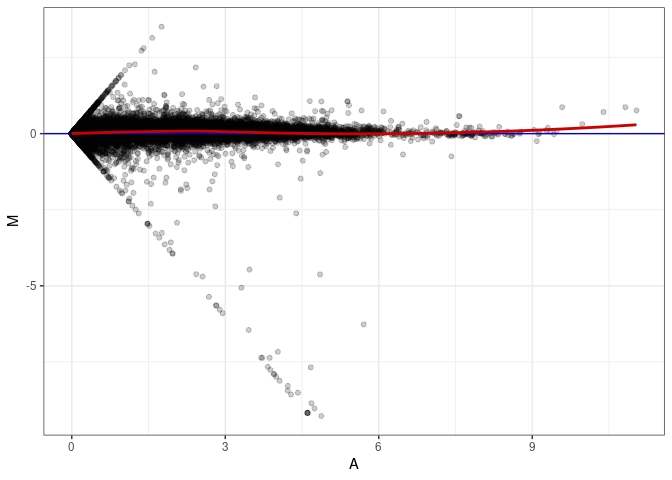
MA plot A064 - A064
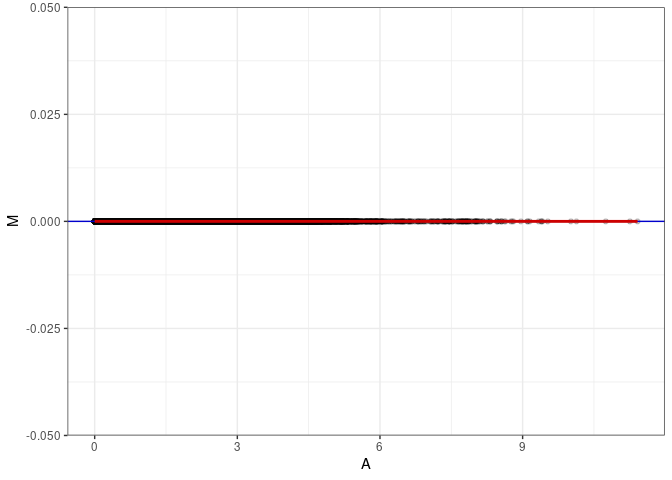
MA plot A064 - A073
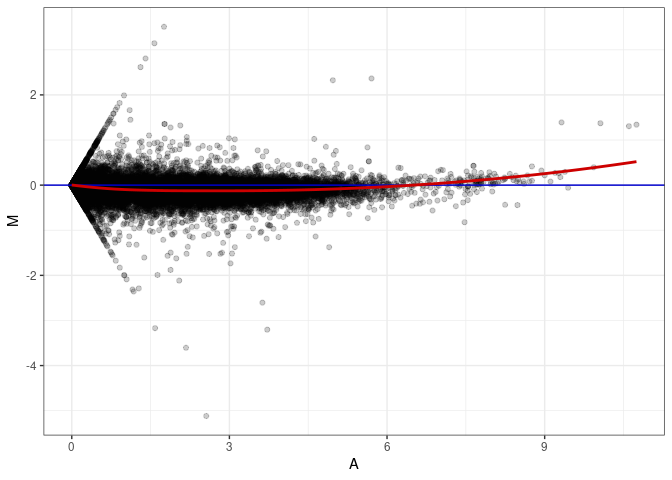
MA plot A064 - A109
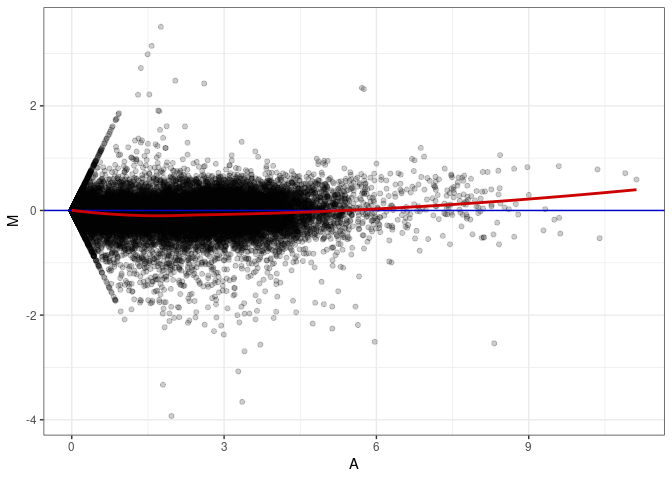
MA plot A073 - A055
MA plot A073 - A064
MA plot A073 - A073
MA plot A073 - A109
MA plot A109 - A055
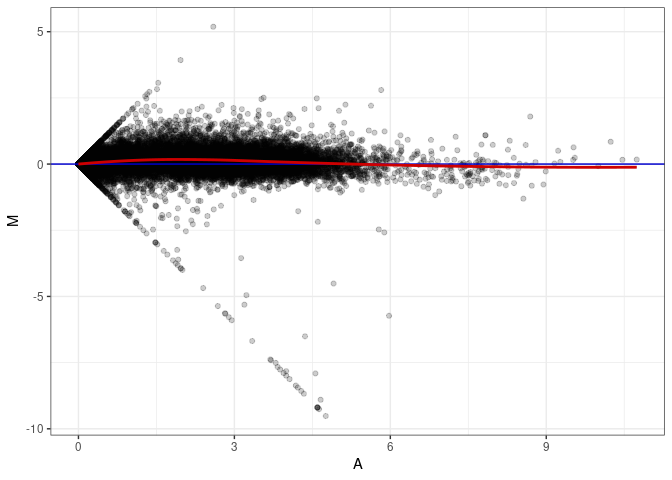
MA plot A109 - A064
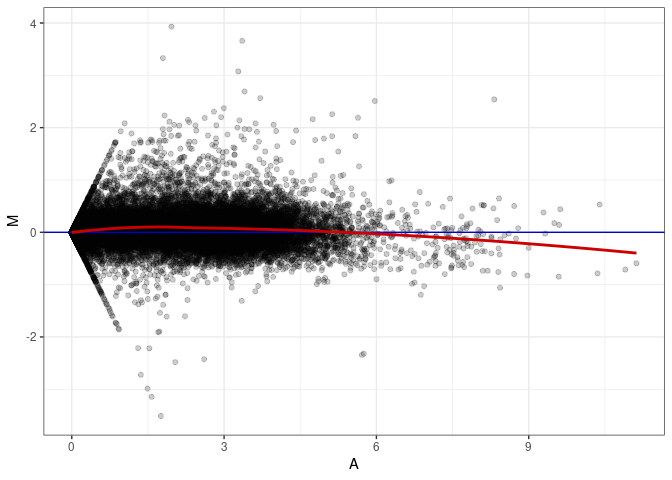
MA plot A109 - A073
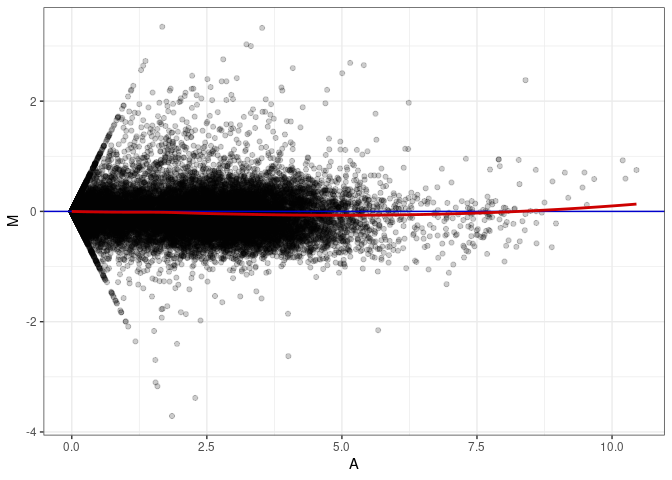
MA plot A109 - A109
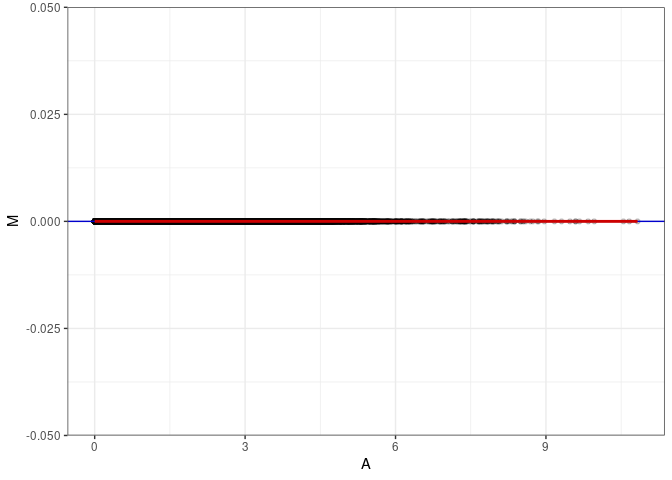
MA plot for Kidney 2DG 4wks
subdata <- log.tdata.FPKM.sample.info %>% rownames_to_column() %>% filter(Tissue =="Kidney" & Treatment == "2DG" & Time == "4 wks") %>% column_to_rownames()
subdata <- subdata[,-c(27237:27239)]
log.data.FPKM.sample.info <- t(subdata)
for(i in 1:ncol(log.data.FPKM.sample.info)){
for(j in 1:ncol(log.data.FPKM.sample.info)){
x = log.data.FPKM.sample.info[,i]
y = log.data.FPKM.sample.info[,j]
## M-values
M=x-y ## A-values
A = (x + y)/2
df = data.frame(A, M)
cat('###',"MA plot", colnames(log.data.FPKM.sample.info)[[i]], "-", colnames(log.data.FPKM.sample.info)[[j]] ,' \n')
p <- ggplot(df, aes(x = A, y = M)) + geom_point(size = 1.5, alpha = 1/5) +
geom_hline(color = "blue3", yintercept = 0) + stat_smooth(se = FALSE, method = "loess", color = "red3") + theme_bw()
print(p)
#print(htmltools::tagList(ggplotly(p)))
cat("\n \n")
}
}MA plot A020 - A020
MA plot A020 - A029
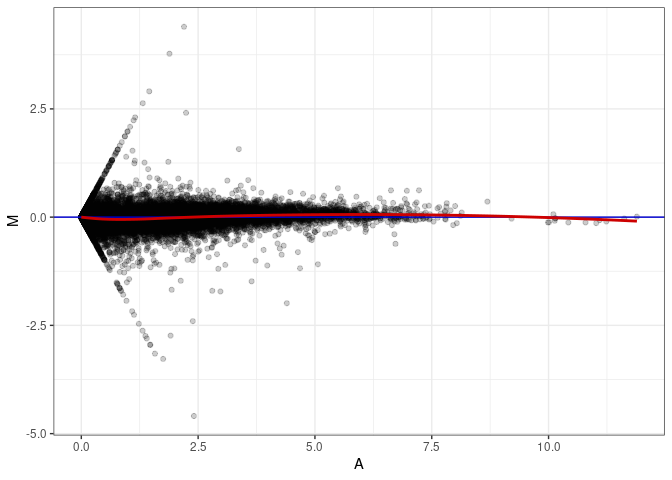
MA plot A020 - A092
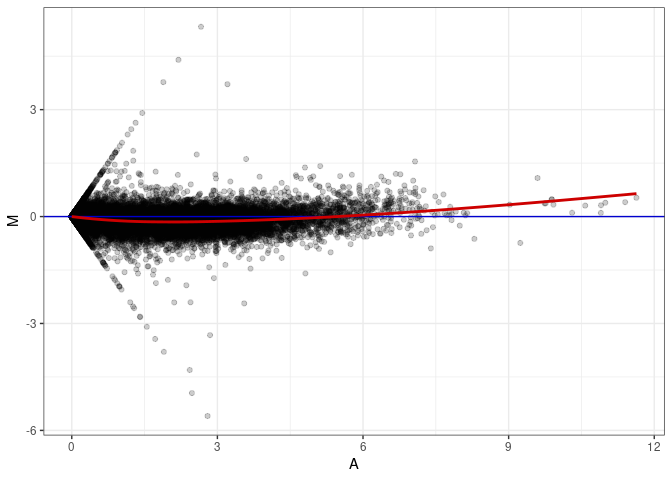
MA plot A020 - A173
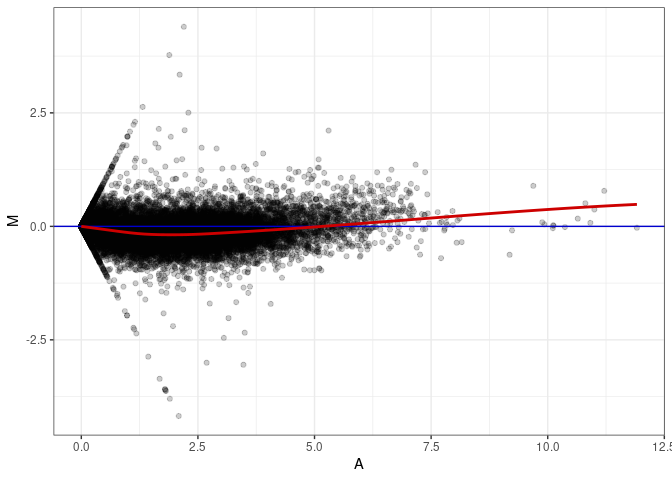
MA plot A029 - A020
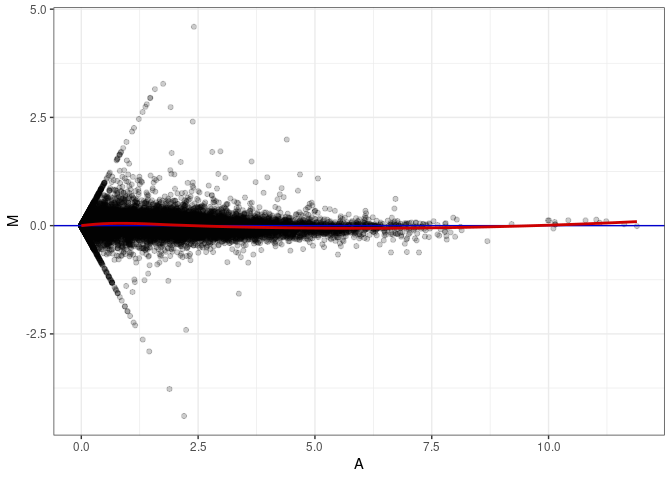
MA plot A029 - A029
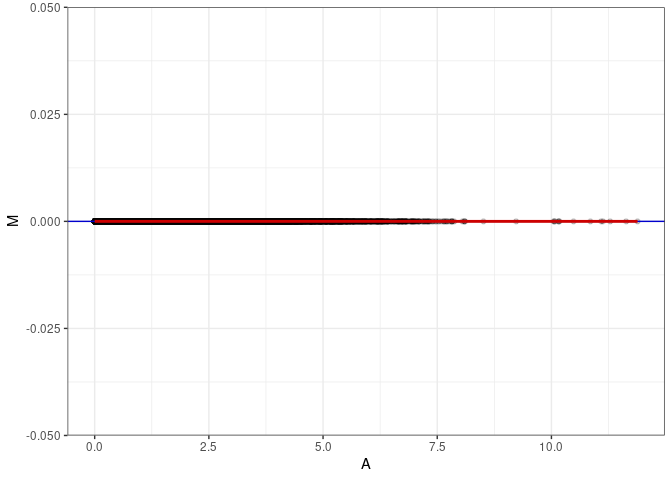
MA plot A029 - A092
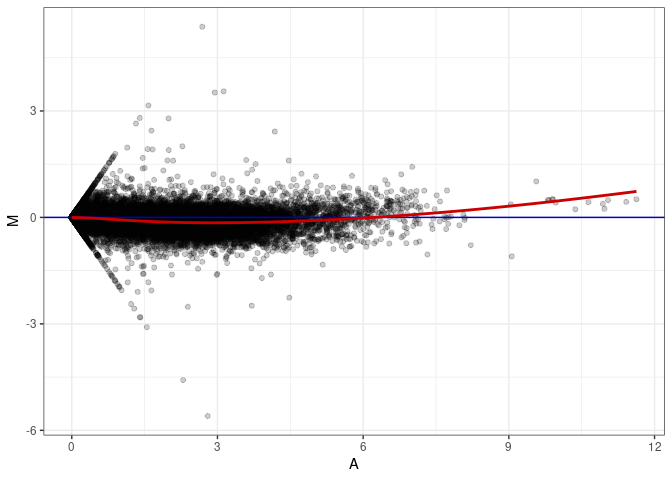
MA plot A029 - A173
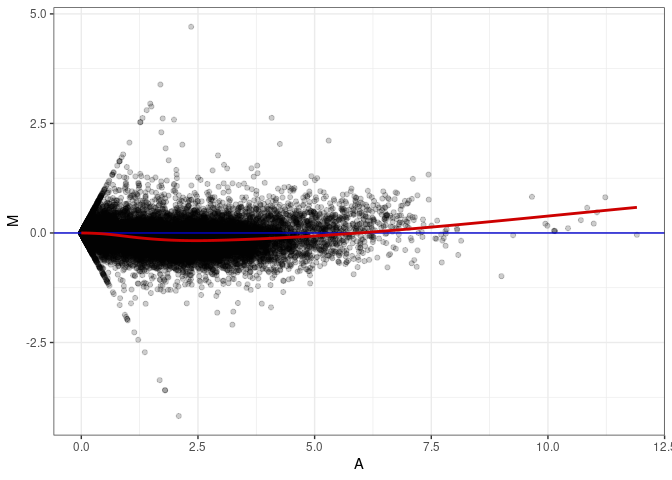
MA plot A092 - A020
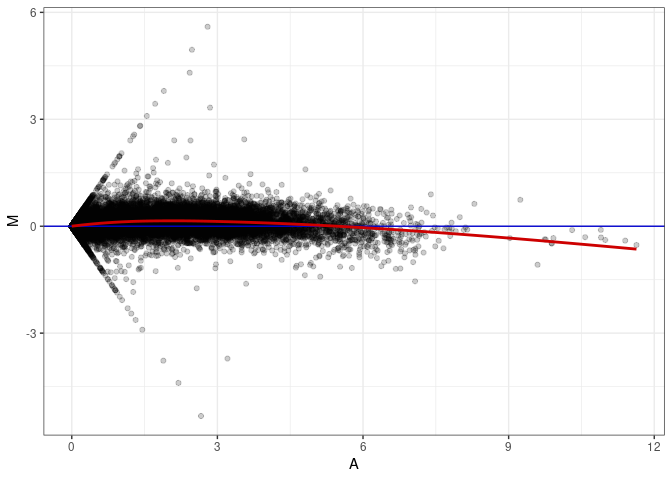
MA plot A092 - A029
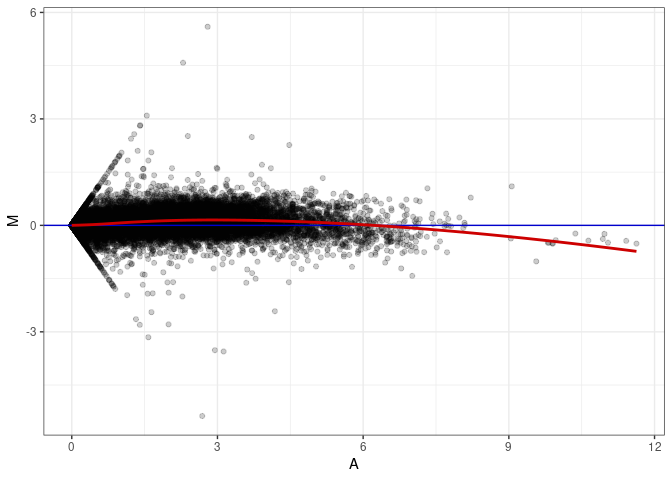
MA plot A092 - A092
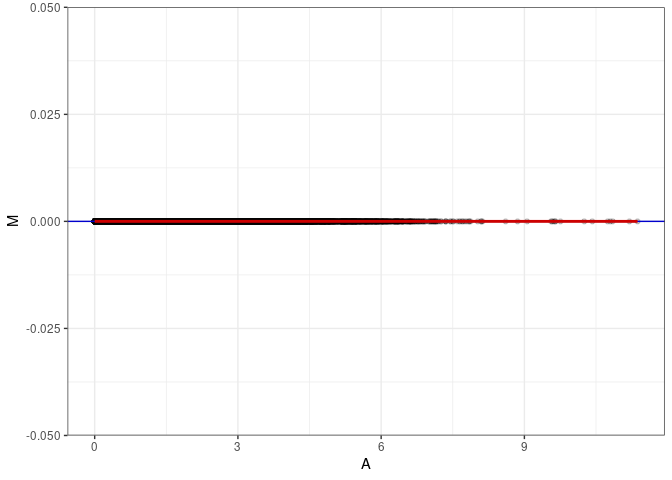
MA plot A092 - A173
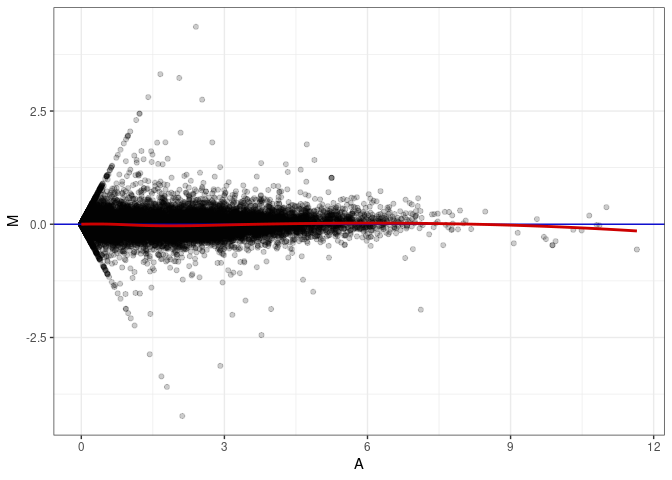
MA plot A173 - A020
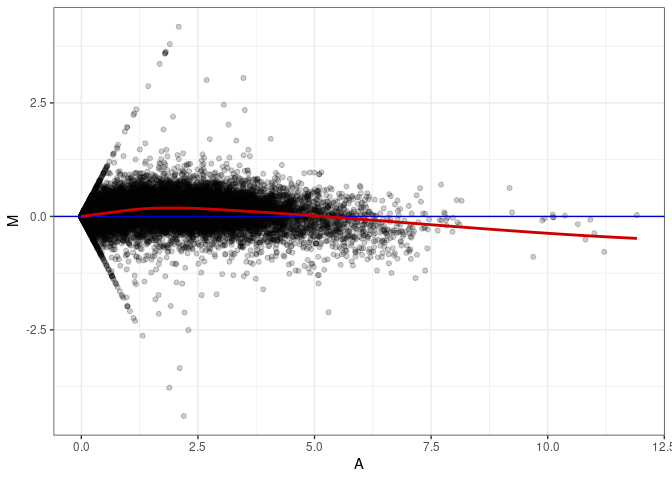
MA plot A173 - A029
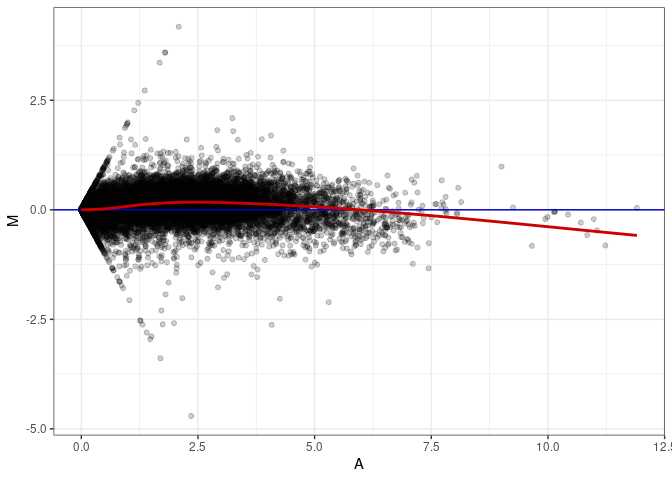
MA plot A173 - A092
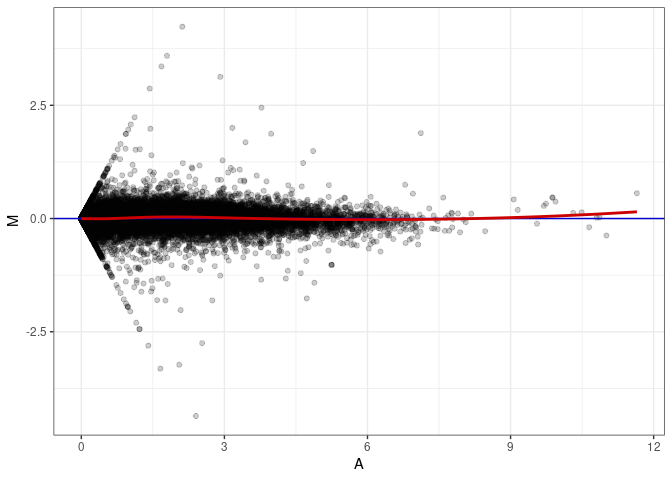
MA plot A173 - A173
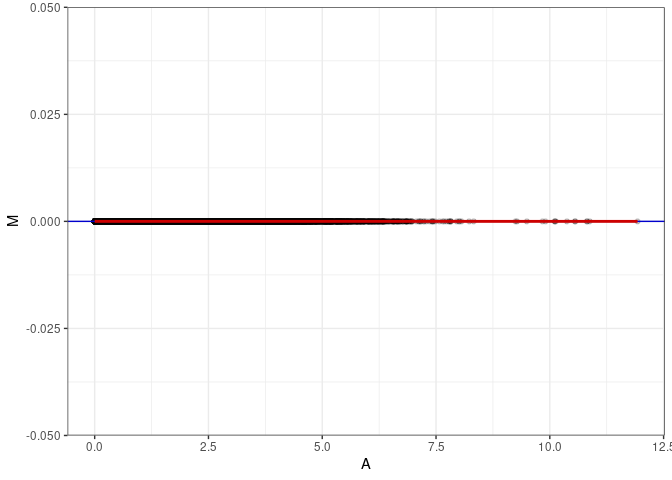
MA plot for Kidney control 4wks
subdata <- log.tdata.FPKM.sample.info %>% rownames_to_column() %>% filter(Tissue =="Kidney" & Treatment == "None" & Time == "4 wks") %>% column_to_rownames()
subdata <- subdata[,-c(27237:27239)]
log.data.FPKM.sample.info <- t(subdata)
for(i in 1:ncol(log.data.FPKM.sample.info)){
for(j in 1:ncol(log.data.FPKM.sample.info)){
x = log.data.FPKM.sample.info[,i]
y = log.data.FPKM.sample.info[,j]
## M-values
M=x-y ## A-values
A = (x + y)/2
df = data.frame(A, M)
cat('###',"MA plot", colnames(log.data.FPKM.sample.info)[[i]], "-", colnames(log.data.FPKM.sample.info)[[j]] ,' \n')
p <- ggplot(df, aes(x = A, y = M)) + geom_point(size = 1.5, alpha = 1/5) +
geom_hline(color = "blue3", yintercept = 0) + stat_smooth(se = FALSE, method = "loess", color = "red3") + theme_bw()
print(p)
#print(htmltools::tagList(ggplotly(p)))
cat("\n \n")
}
}MA plot A002 - A002
MA plot A002 - A038
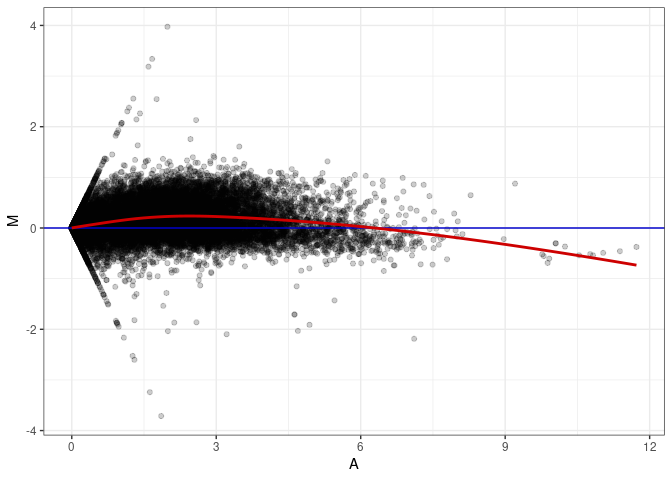
MA plot A002 - A047
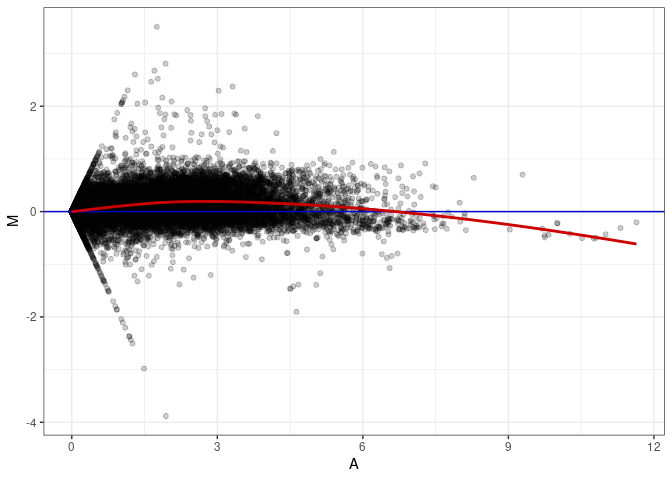
MA plot A002 - A083
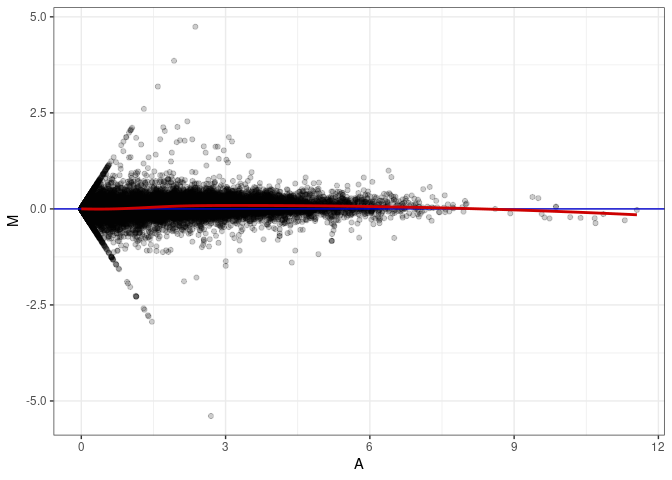
MA plot A038 - A002
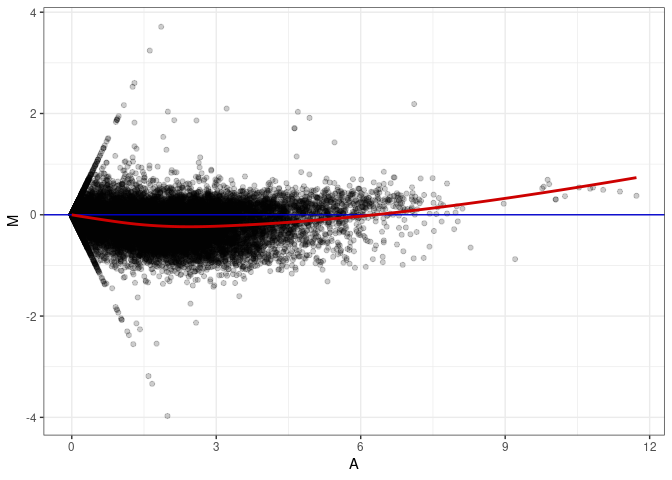
MA plot A038 - A038
MA plot A038 - A047
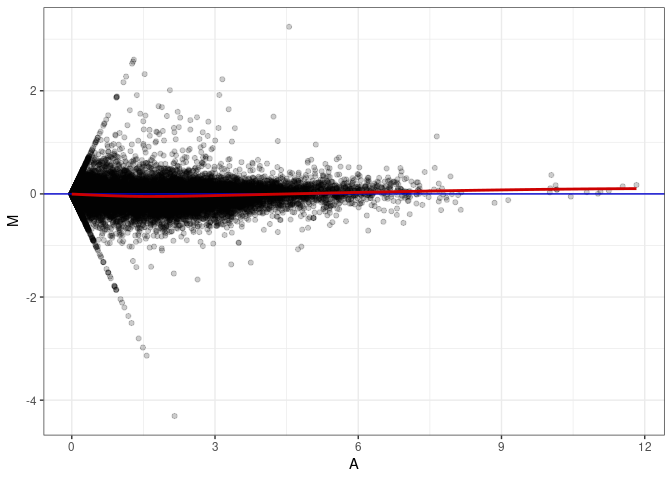
MA plot A038 - A083
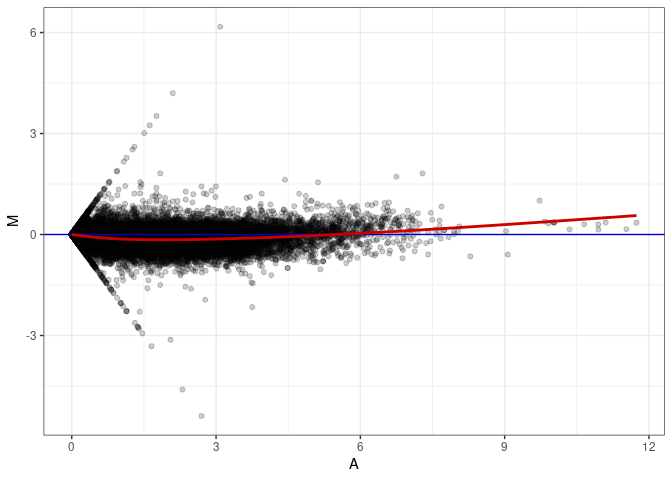
MA plot A047 - A002
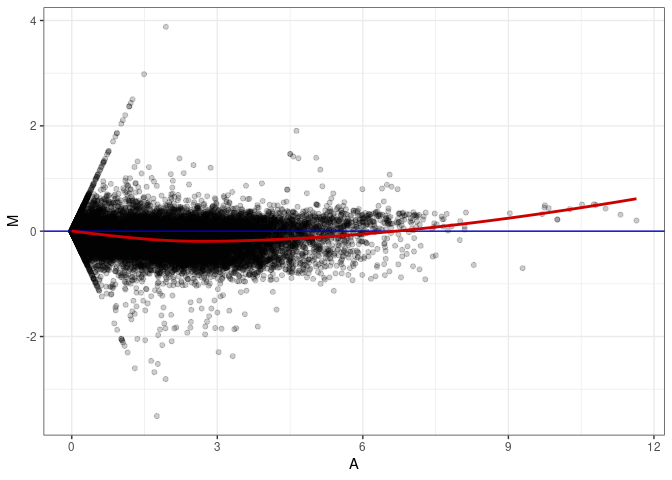
MA plot A047 - A038
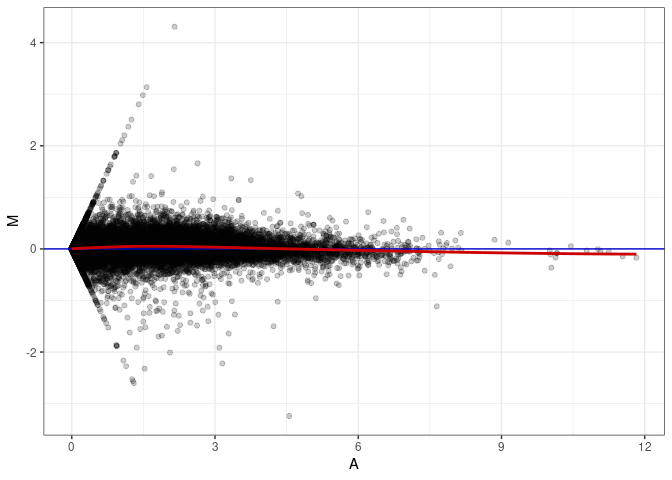
MA plot A047 - A047
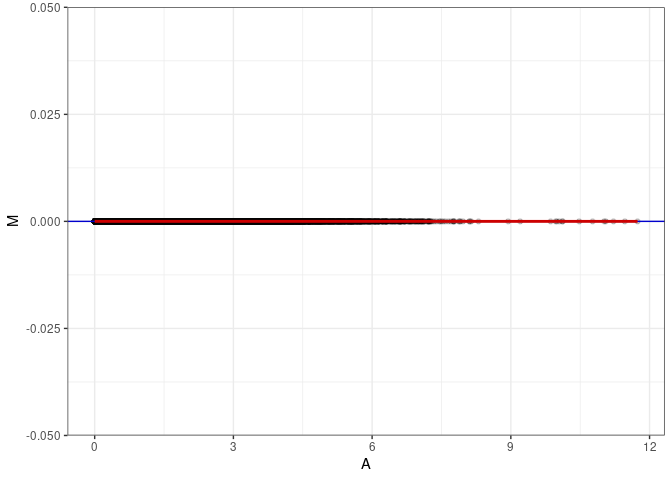
MA plot A047 - A083
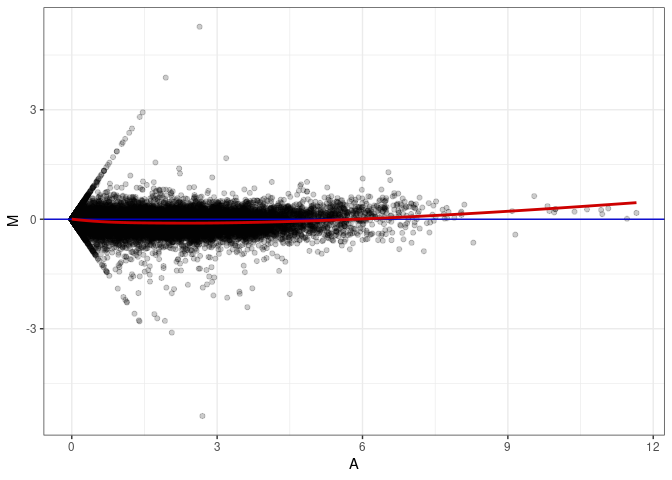
MA plot A083 - A002
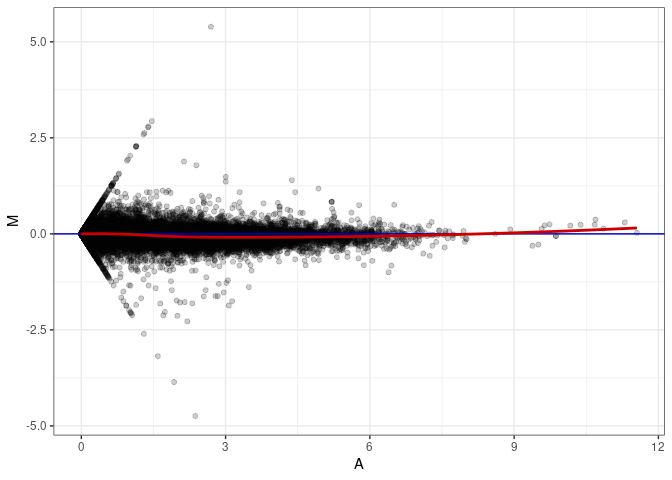
MA plot A083 - A038
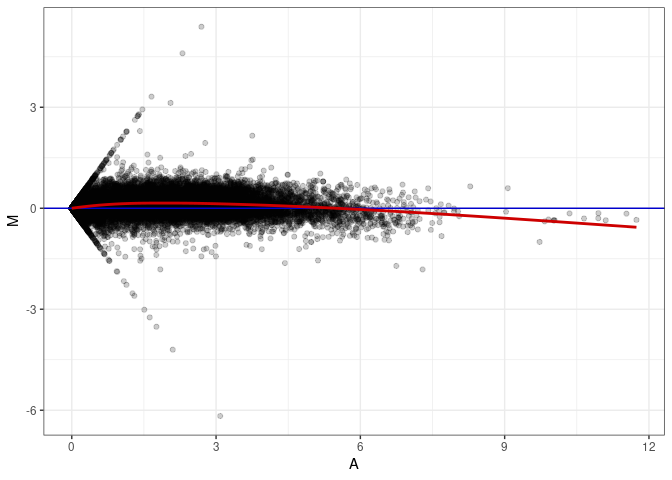
MA plot A083 - A047
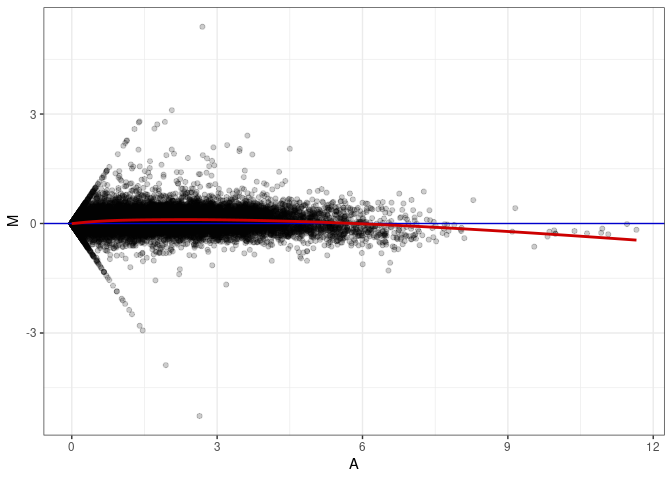
MA plot A083 - A083
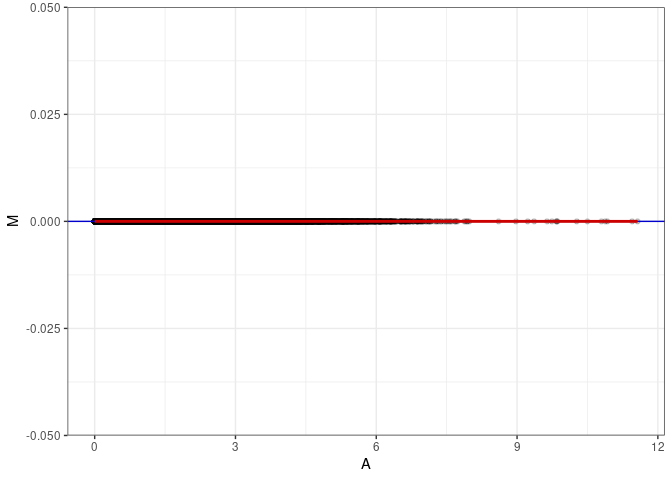
MA plot for Kidney 2DG 96hrs
subdata <- log.tdata.FPKM.sample.info %>% rownames_to_column() %>% filter(Tissue =="Kidney" & Treatment == "2DG" & Time == "96 hrs") %>% column_to_rownames()
subdata <- subdata[,-c(27237:27239)]
log.data.FPKM.sample.info <- t(subdata)
for(i in 1:ncol(log.data.FPKM.sample.info)){
for(j in 1:ncol(log.data.FPKM.sample.info)){
x = log.data.FPKM.sample.info[,i]
y = log.data.FPKM.sample.info[,j]
## M-values
M=x-y ## A-values
A = (x + y)/2
df = data.frame(A, M)
cat('###',"MA plot", colnames(log.data.FPKM.sample.info)[[i]], "-", colnames(log.data.FPKM.sample.info)[[j]] ,' \n')
p <- ggplot(df, aes(x = A, y = M)) + geom_point(size = 1.5, alpha = 1/5) +
geom_hline(color = "blue3", yintercept = 0) + stat_smooth(se = FALSE, method = "loess", color = "red3") + theme_bw()
print(p)
#print(htmltools::tagList(ggplotly(p)))
cat("\n \n")
}
}MA plot A011 - A011
MA plot A011 - A137
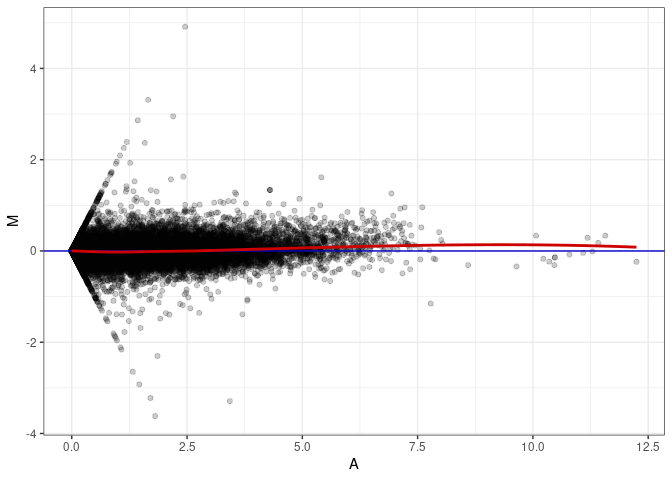
MA plot A011 - A146
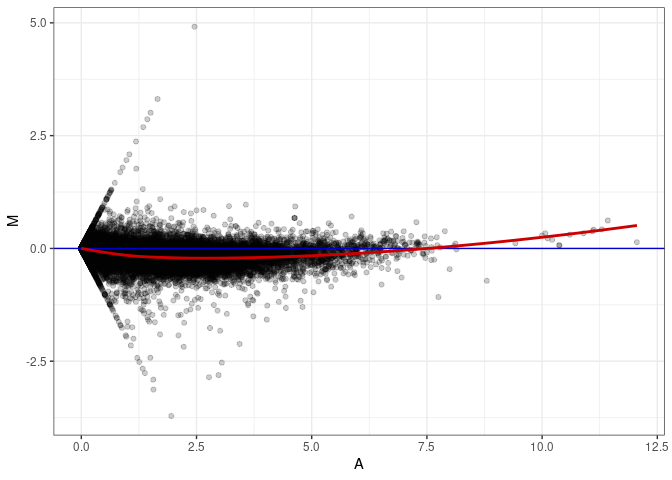
MA plot A011 - A164
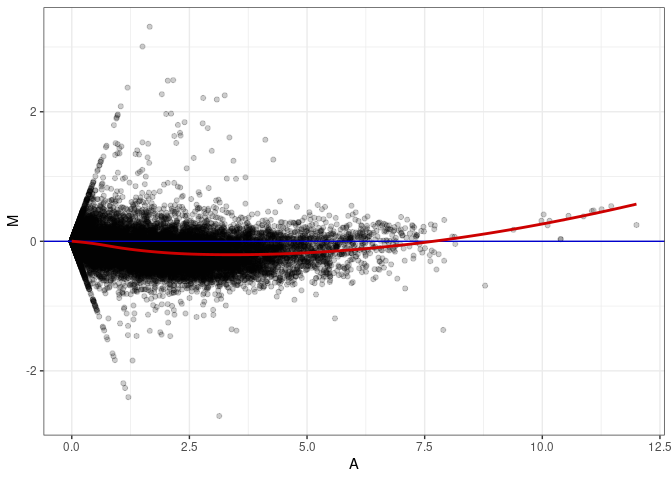
MA plot A137 - A011
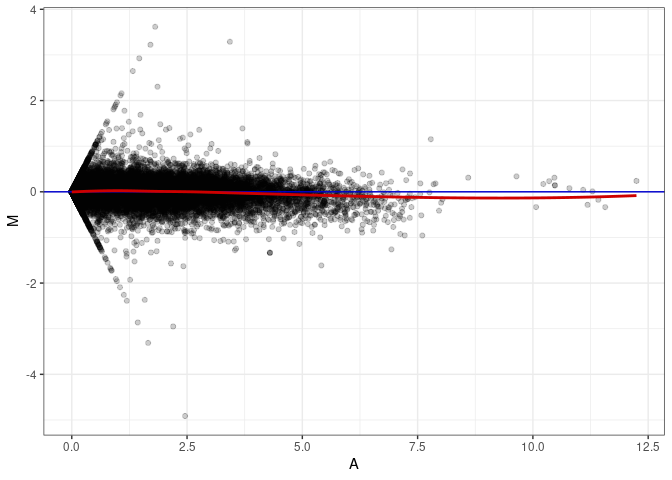
MA plot A137 - A137
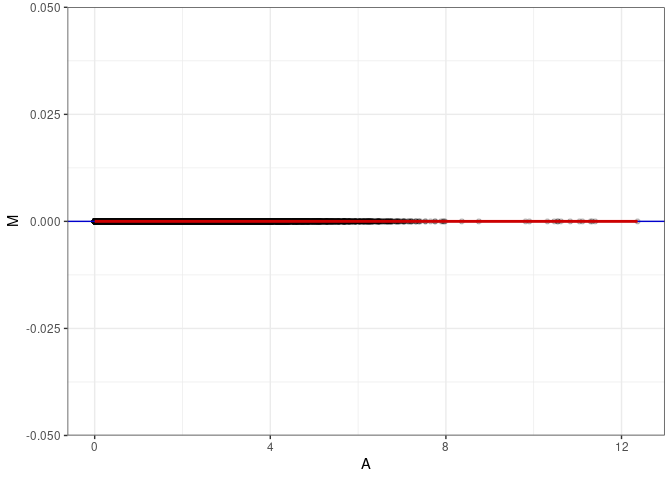
MA plot A137 - A146
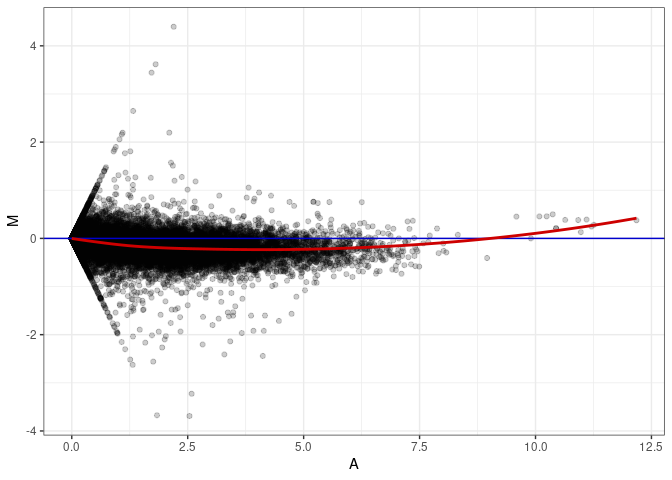
MA plot A137 - A164
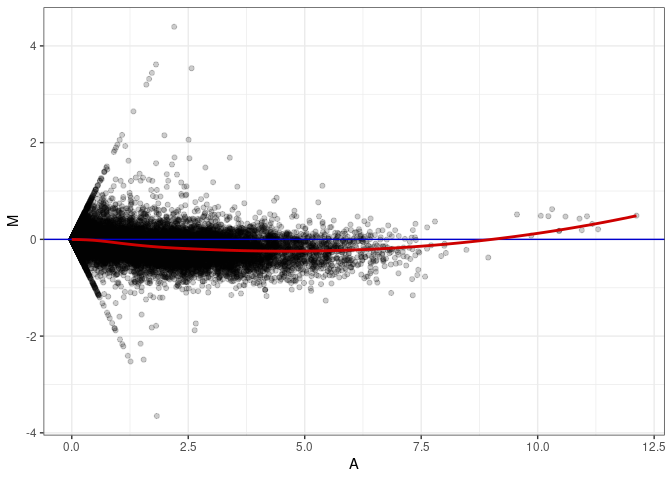
MA plot A146 - A011
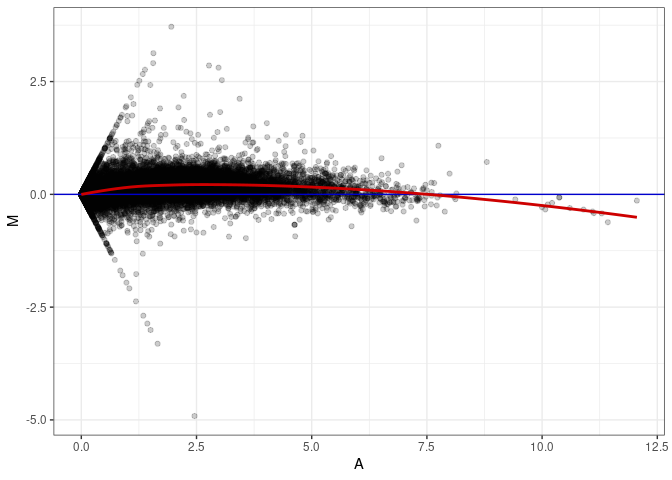
MA plot A146 - A137
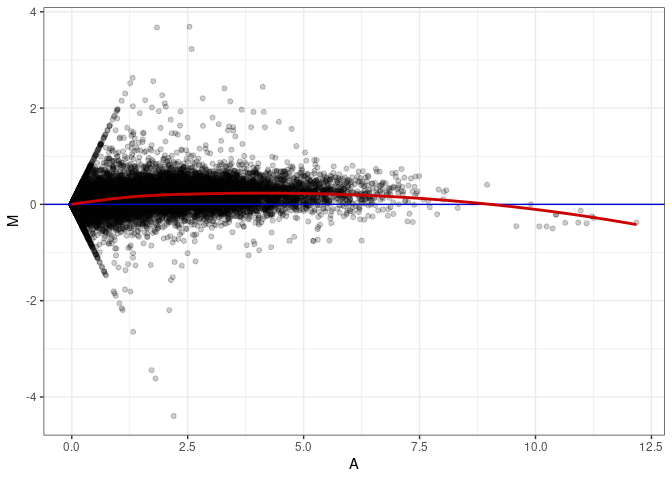
MA plot A146 - A146
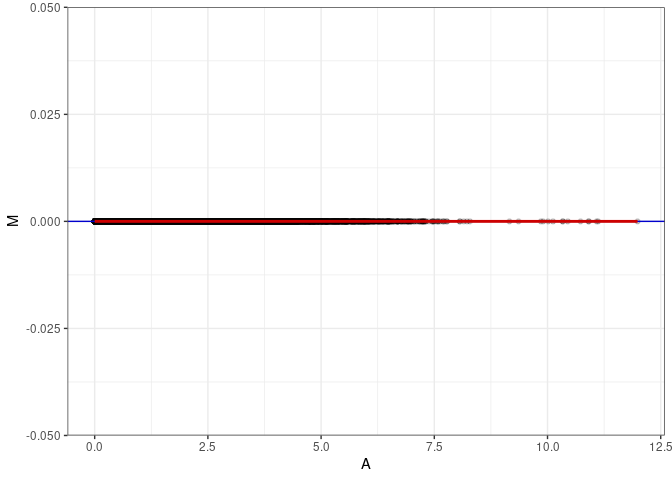
MA plot A146 - A164
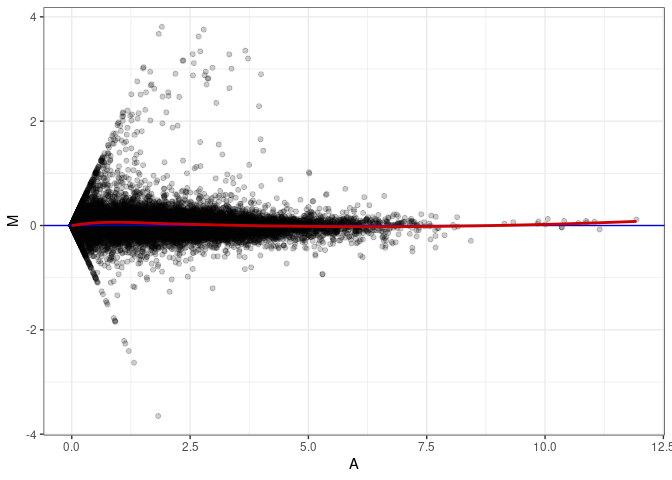
MA plot A164 - A011
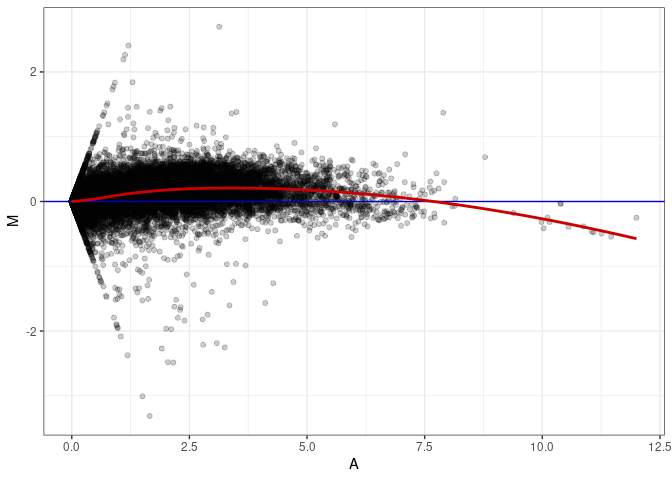
MA plot A164 - A137
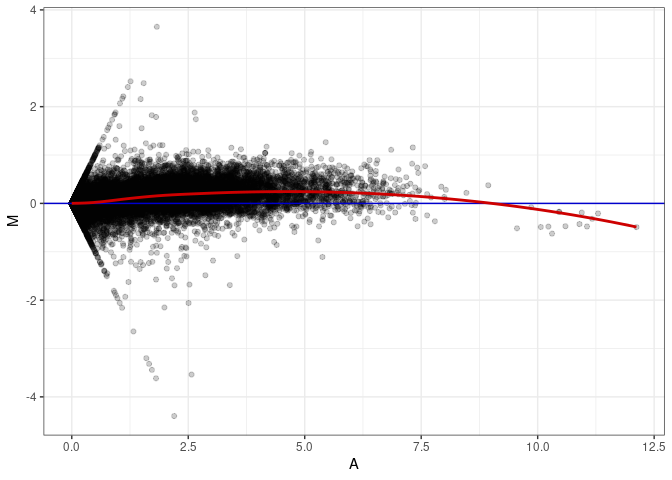
MA plot A164 - A146
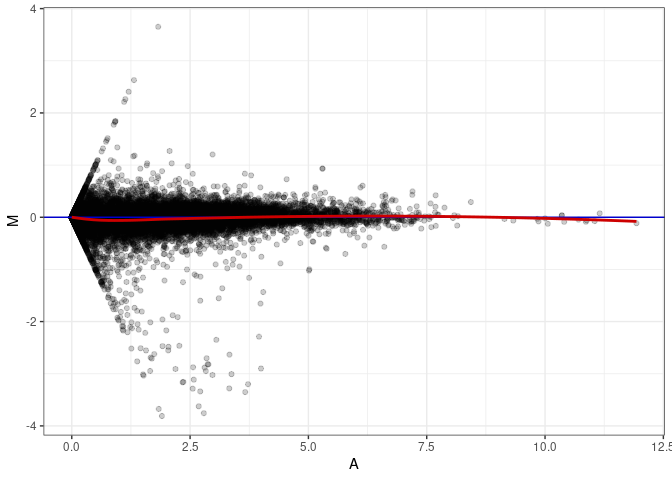
MA plot A164 - A164
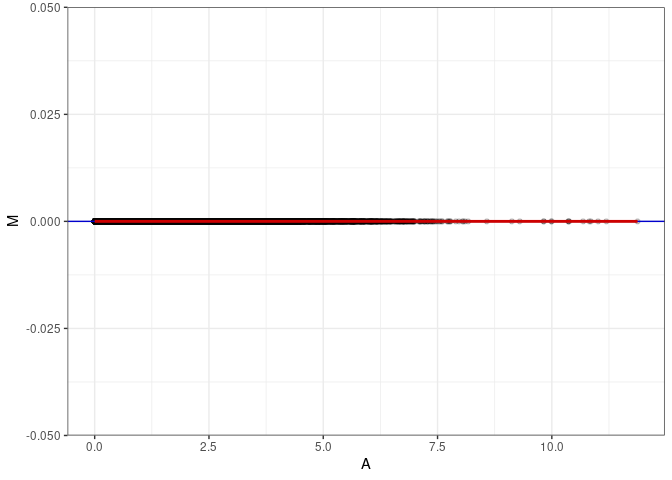
MA plot for Kidney control 96hrs
subdata <- log.tdata.FPKM.sample.info %>% rownames_to_column() %>% filter(Tissue =="Kidney" & Treatment == "None" & Time == "96 hrs") %>% column_to_rownames()
subdata <- subdata[,-c(27237:27239)]
log.data.FPKM.sample.info <- t(subdata)
for(i in 1:ncol(log.data.FPKM.sample.info)){
for(j in 1:ncol(log.data.FPKM.sample.info)){
x = log.data.FPKM.sample.info[,i]
y = log.data.FPKM.sample.info[,j]
## M-values
M=x-y ## A-values
A = (x + y)/2
df = data.frame(A, M)
cat('###',"MA plot", colnames(log.data.FPKM.sample.info)[[i]], "-", colnames(log.data.FPKM.sample.info)[[j]] ,' \n')
p <- ggplot(df, aes(x = A, y = M)) + geom_point(size = 1.5, alpha = 1/5) +
geom_hline(color = "blue3", yintercept = 0) + stat_smooth(se = FALSE, method = "loess", color = "red3") + theme_bw()
print(p)
#print(htmltools::tagList(ggplotly(p)))
cat("\n \n")
}
}MA plot A056 - A056
MA plot A056 - A065
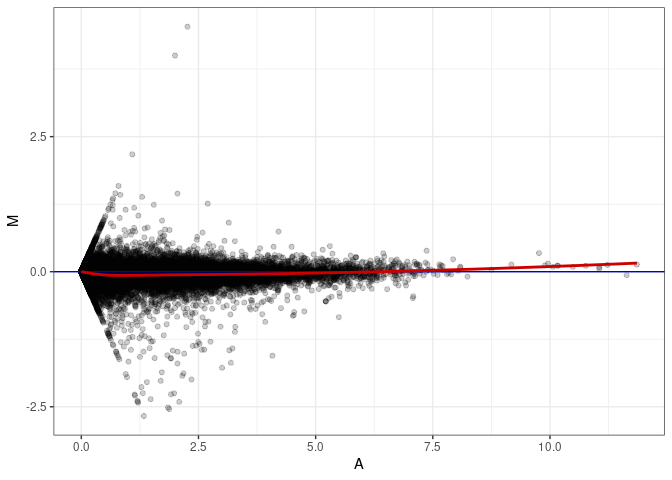
MA plot A056 - A074
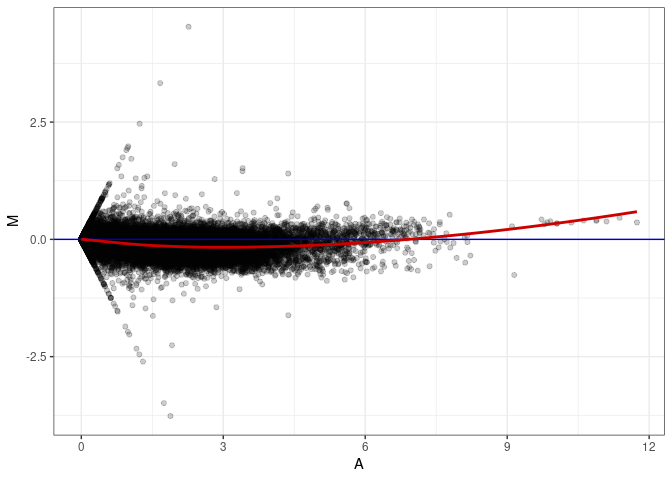
MA plot A056 - A110
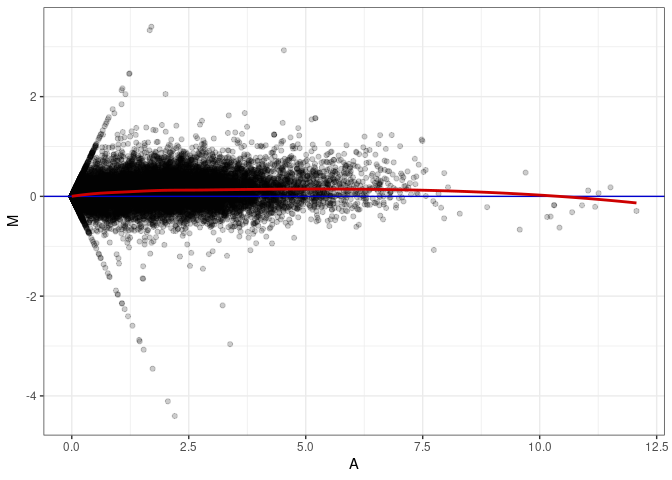
MA plot A065 - A056
MA plot A065 - A065
MA plot A065 - A074
MA plot A065 - A110
MA plot A074 - A056
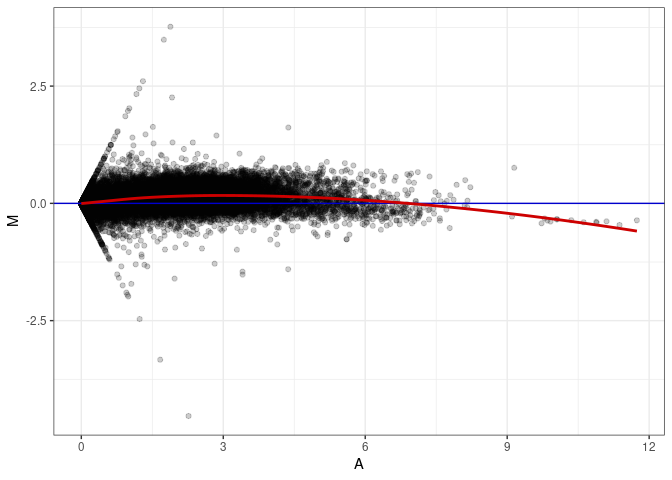
MA plot A074 - A065
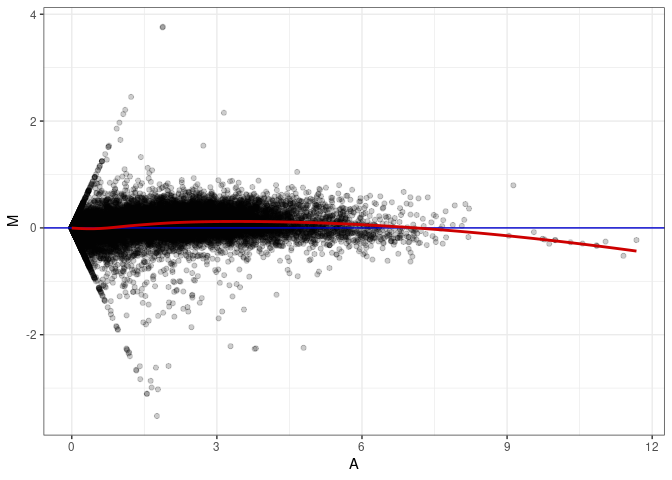
MA plot A074 - A074
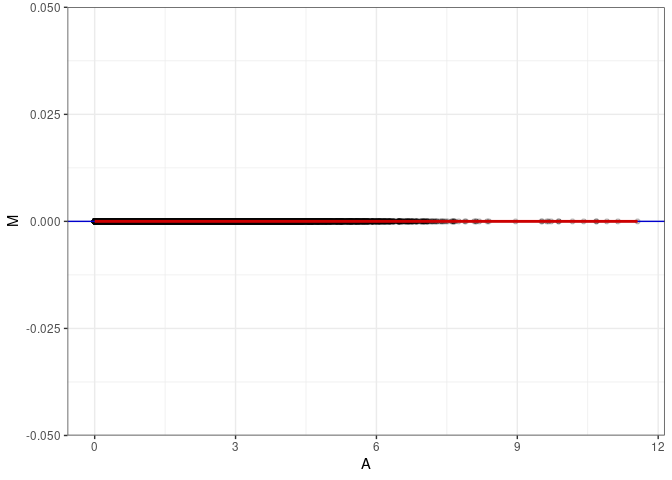
MA plot A074 - A110
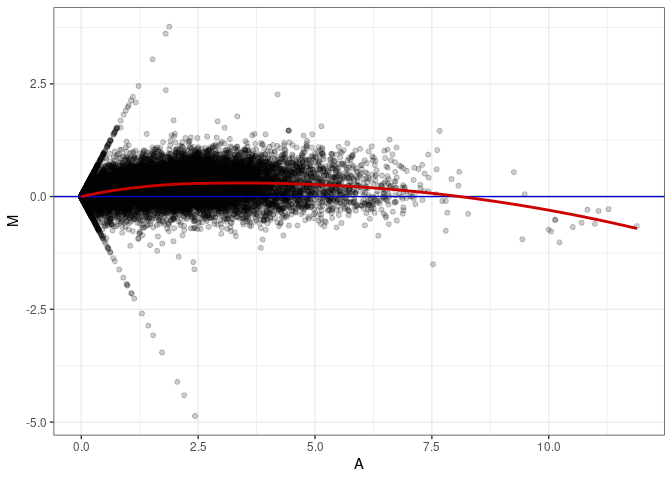
MA plot A110 - A056
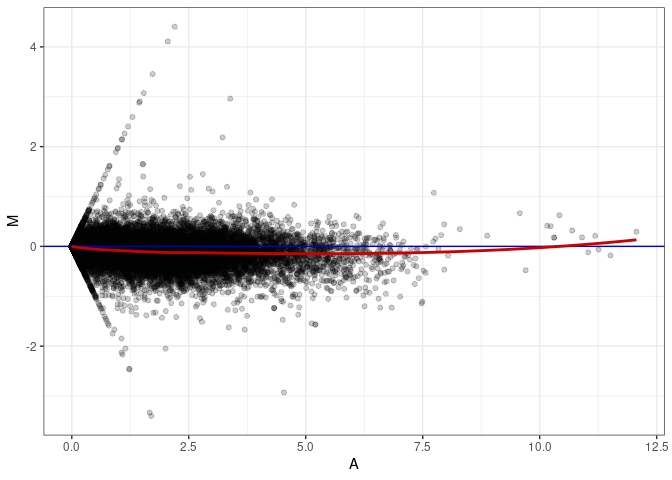
MA plot A110 - A065
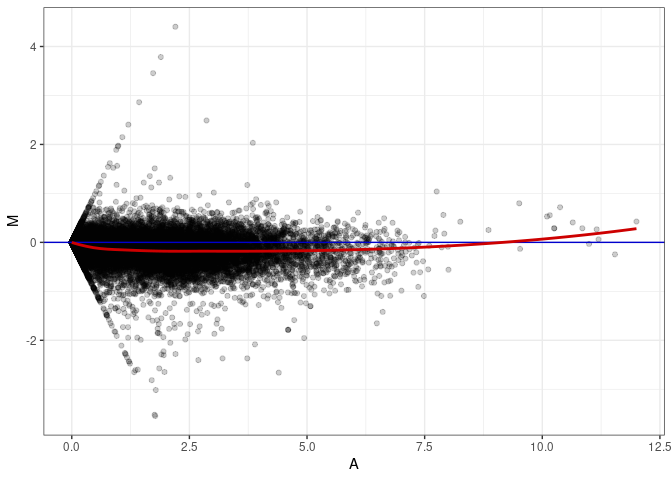
MA plot A110 - A074
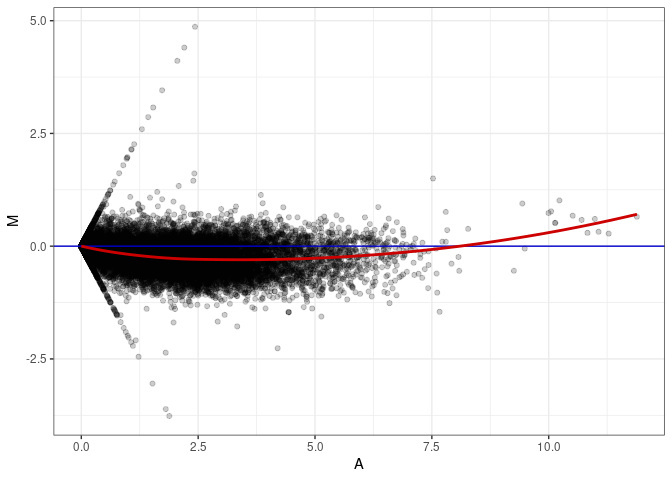
MA plot A110 - A110
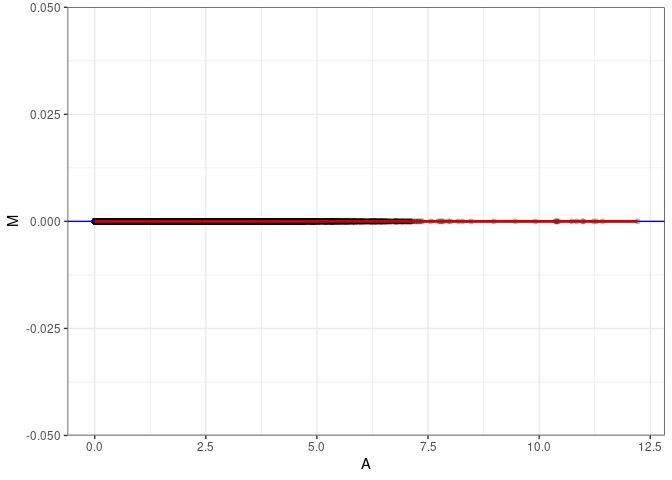
MA plot for Hypothalamus 2DG 4wks
subdata <- log.tdata.FPKM.sample.info %>% rownames_to_column() %>% filter(Tissue =="Hypothanamus" & Treatment == "2DG" & Time == "4 wks") %>% column_to_rownames()
subdata <- subdata[,-c(27237:27239)]
log.data.FPKM.sample.info <- t(subdata)
for(i in 1:ncol(log.data.FPKM.sample.info)){
for(j in 1:ncol(log.data.FPKM.sample.info)){
x = log.data.FPKM.sample.info[,i]
y = log.data.FPKM.sample.info[,j]
## M-values
M=x-y ## A-values
A = (x + y)/2
df = data.frame(A, M)
cat('###',"MA plot", colnames(log.data.FPKM.sample.info)[[i]], "-", colnames(log.data.FPKM.sample.info)[[j]] ,' \n')
p <- ggplot(df, aes(x = A, y = M)) + geom_point(size = 1.5, alpha = 1/5) +
geom_hline(color = "blue3", yintercept = 0) + stat_smooth(se = FALSE, method = "loess", color = "red3") + theme_bw()
print(p)
#print(htmltools::tagList(ggplotly(p)))
cat("\n \n")
}
}MA plot A025 - A025
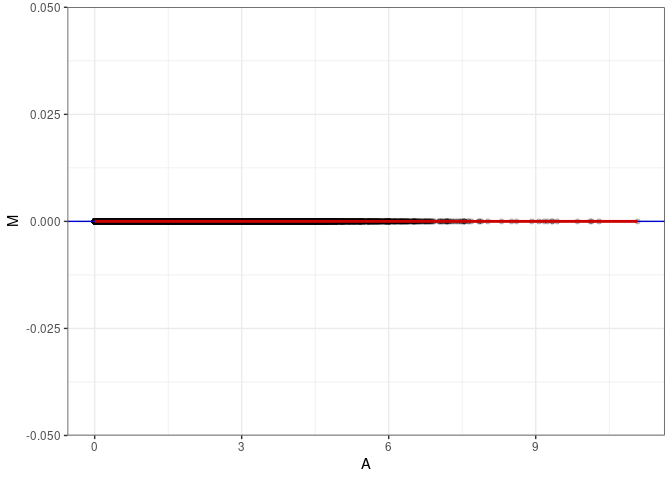
MA plot A025 - A034
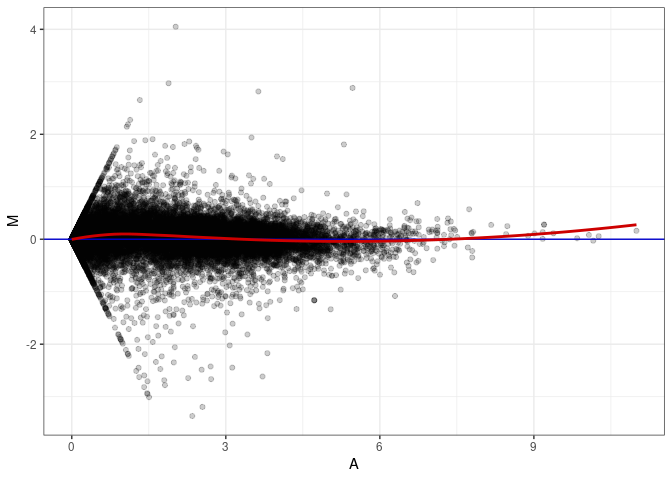
MA plot A025 - A097
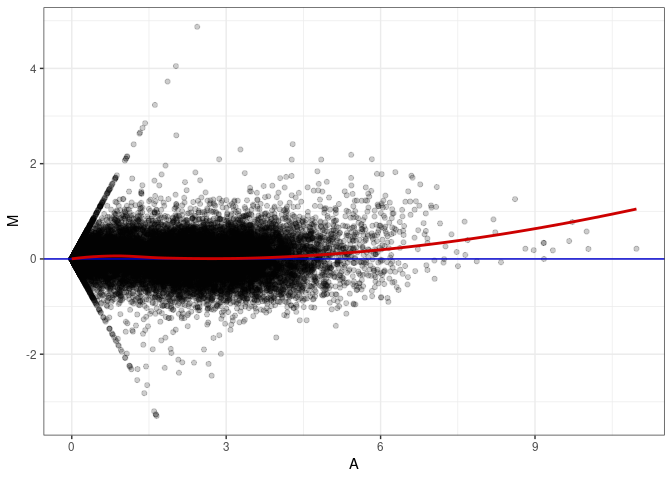
MA plot A025 - A178
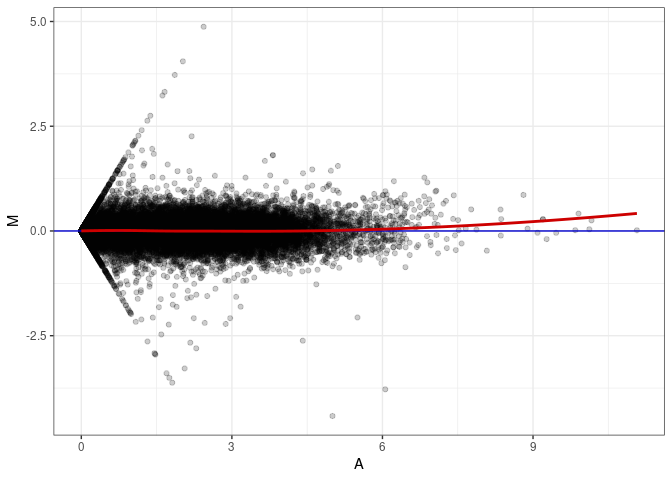
MA plot A034 - A025
MA plot A034 - A034
MA plot A034 - A097
MA plot A034 - A178
MA plot A097 - A025
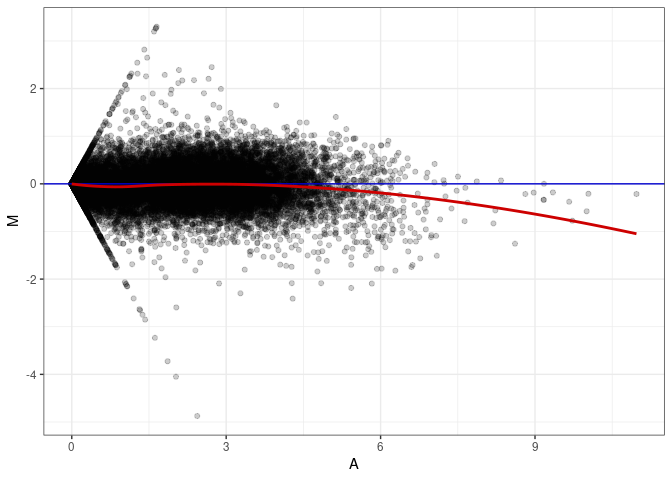
MA plot A097 - A034
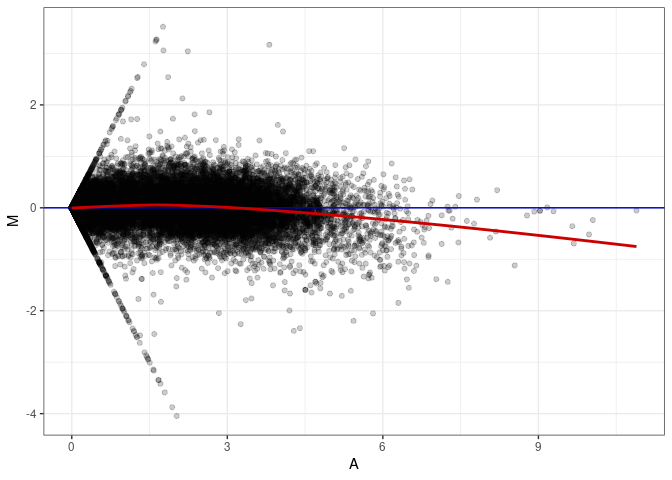
MA plot A097 - A097
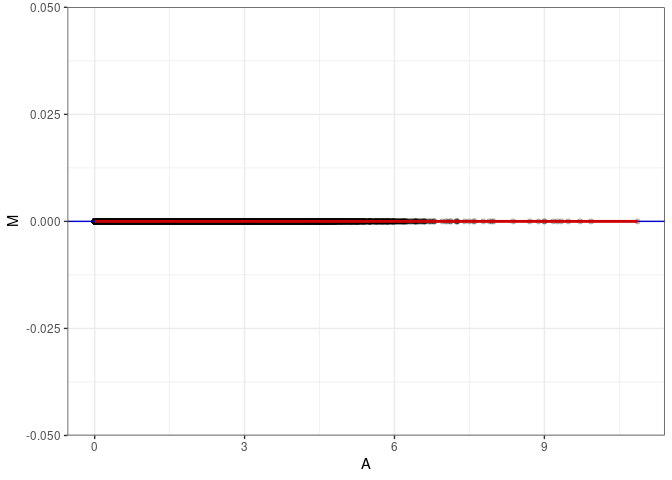
MA plot A097 - A178
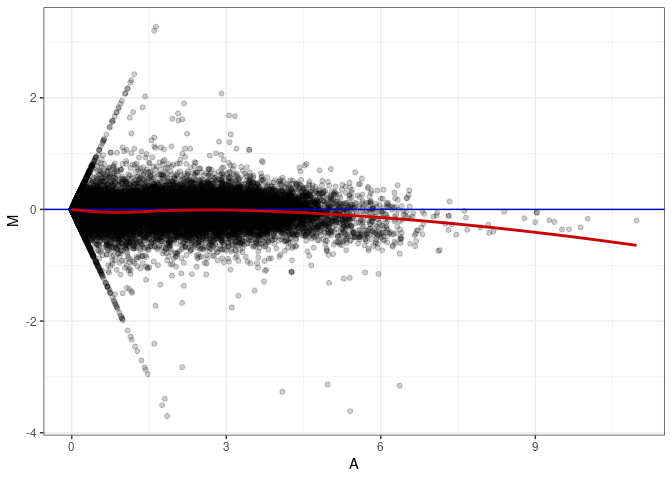
MA plot A178 - A025
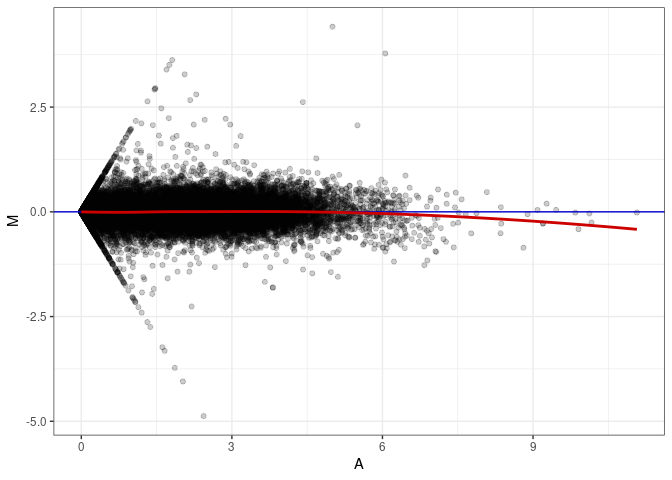
MA plot A178 - A034
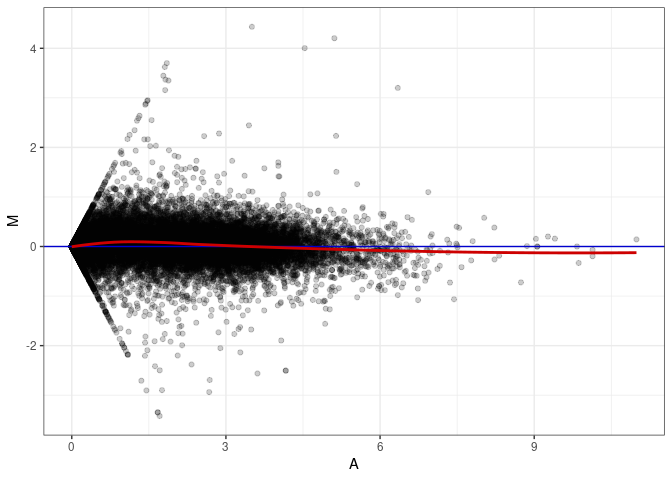
MA plot A178 - A097
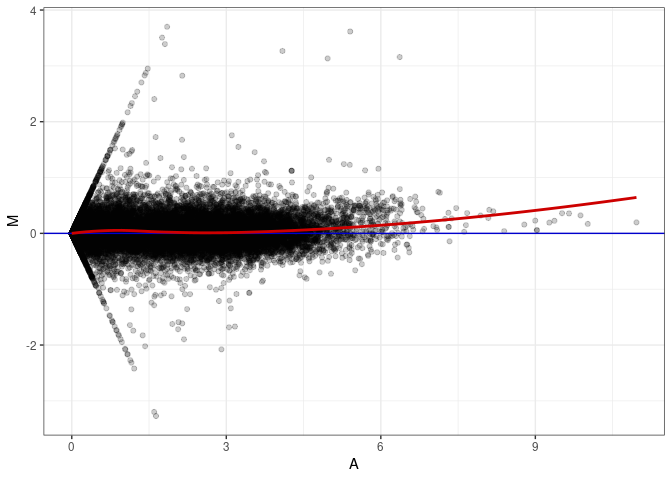
MA plot A178 - A178
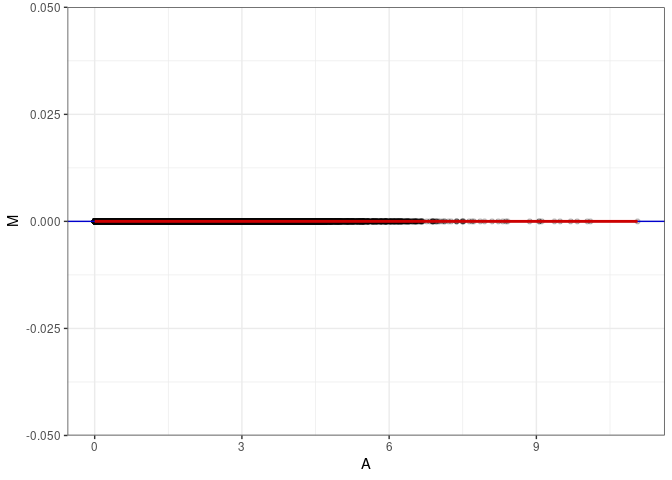
MA plot for Hypothalamus control 4wks
subdata <- log.tdata.FPKM.sample.info %>% rownames_to_column() %>% filter(Tissue =="Hypothanamus" & Treatment == "None" & Time == "4 wks") %>% column_to_rownames()
subdata <- subdata[,-c(27237:27239)]
log.data.FPKM.sample.info <- t(subdata)
for(i in 1:ncol(log.data.FPKM.sample.info)){
for(j in 1:ncol(log.data.FPKM.sample.info)){
x = log.data.FPKM.sample.info[,i]
y = log.data.FPKM.sample.info[,j]
## M-values
M=x-y ## A-values
A = (x + y)/2
df = data.frame(A, M)
cat('###',"MA plot", colnames(log.data.FPKM.sample.info)[[i]], "-", colnames(log.data.FPKM.sample.info)[[j]] ,' \n')
p <- ggplot(df, aes(x = A, y = M)) + geom_point(size = 1.5, alpha = 1/5) +
geom_hline(color = "blue3", yintercept = 0) + stat_smooth(se = FALSE, method = "loess", color = "red3") + theme_bw()
print(p)
#print(htmltools::tagList(ggplotly(p)))
cat("\n \n")
}
}MA plot A007 - A007
MA plot A007 - A043
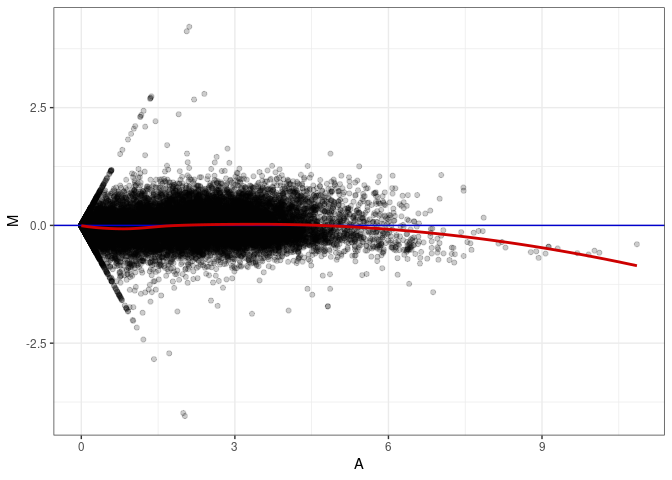
MA plot A007 - A052
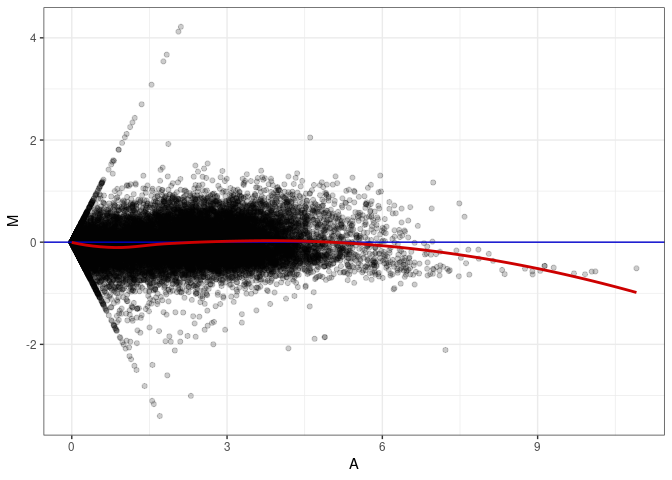
MA plot A007 - A088
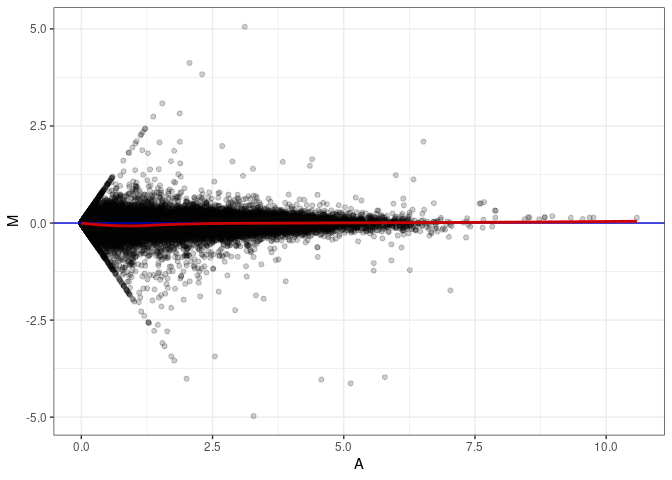
MA plot A043 - A007
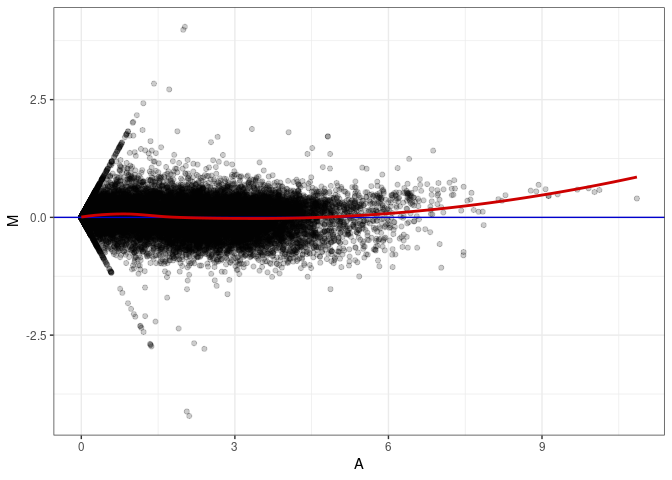
MA plot A043 - A043
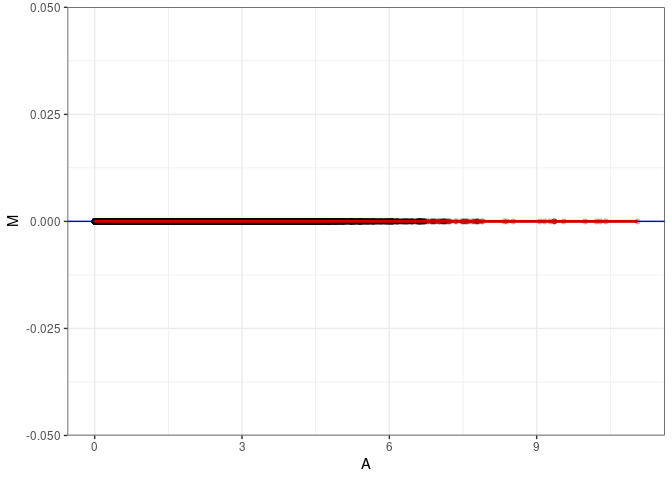
MA plot A043 - A052
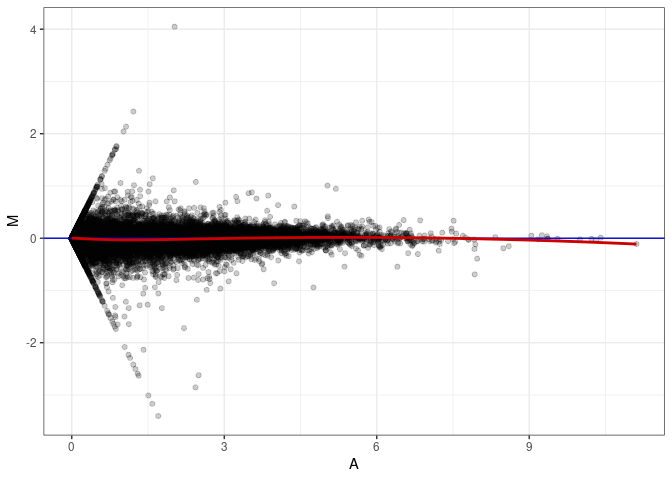
MA plot A043 - A088
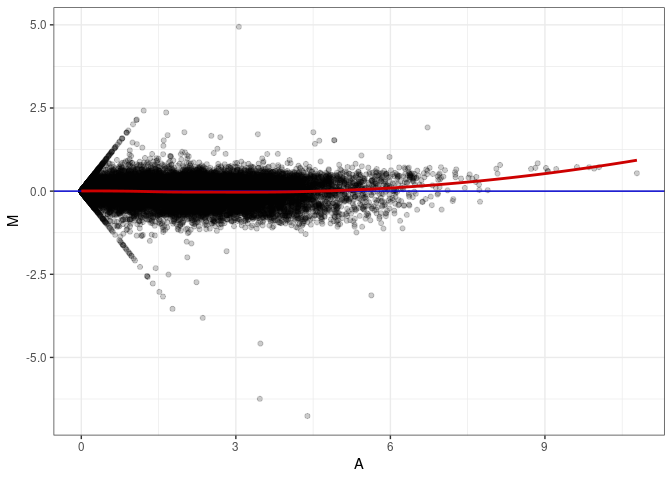
MA plot A052 - A007
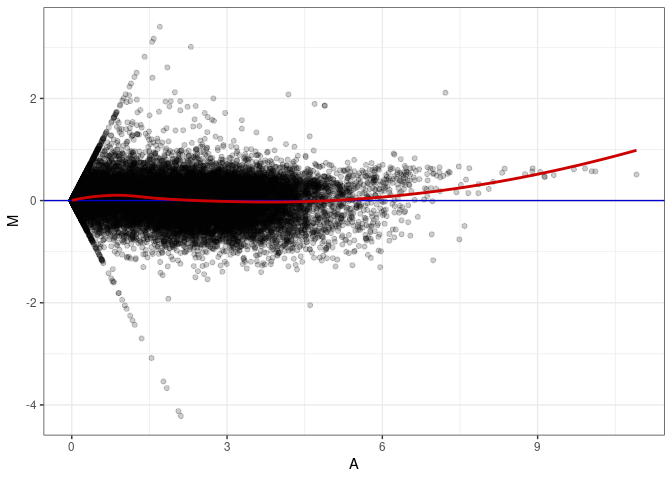
MA plot A052 - A043
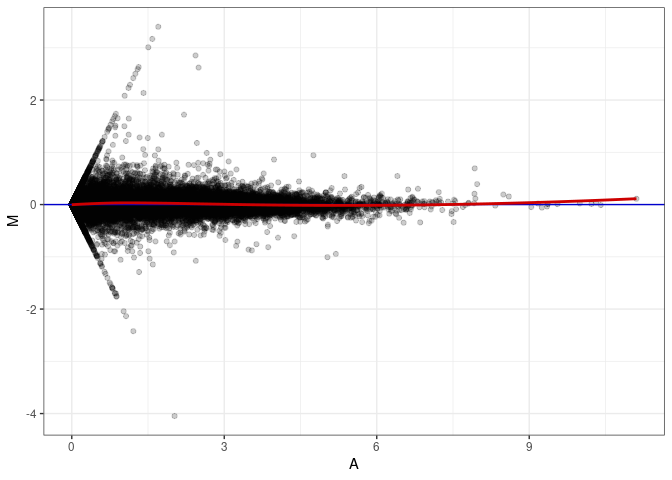
MA plot A052 - A052
MA plot A052 - A088
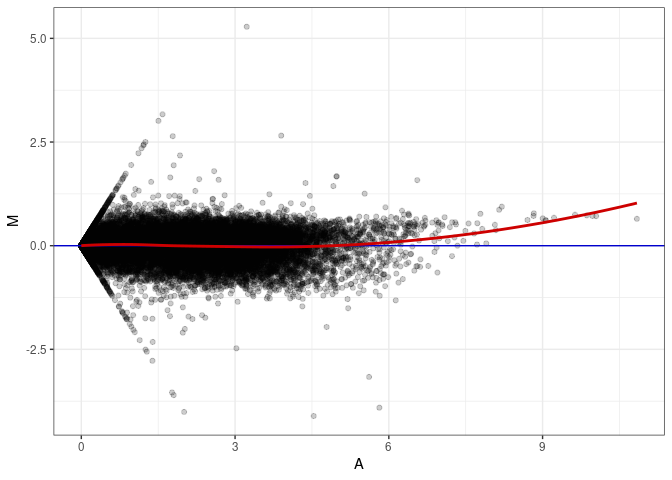
MA plot A088 - A007
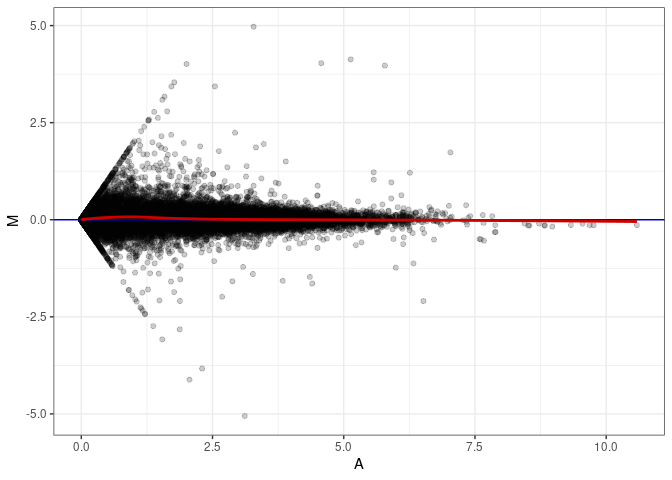
MA plot A088 - A043
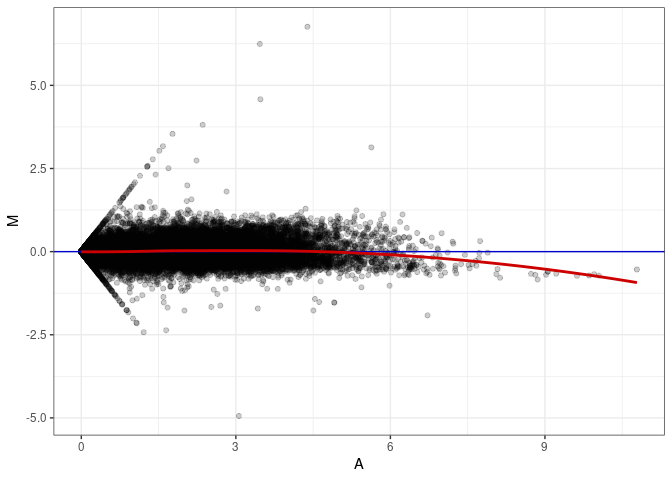
MA plot A088 - A052
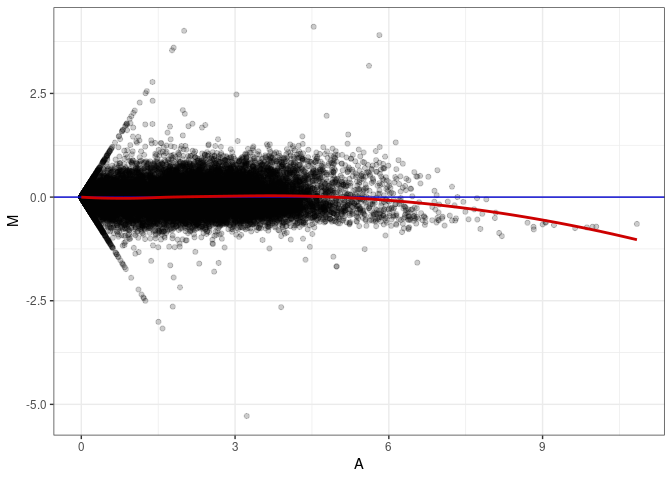
MA plot A088 - A088
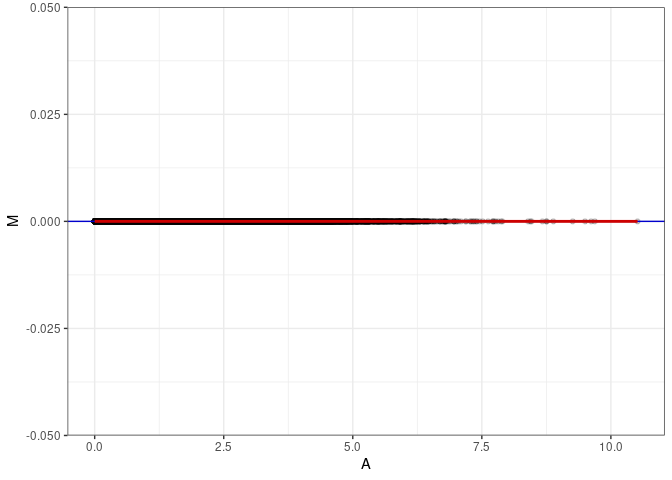
MA plot for Hypothalamus 2DG 96hrs
subdata <- log.tdata.FPKM.sample.info %>% rownames_to_column() %>% filter(Tissue =="Hypothanamus" & Treatment == "2DG" & Time == "96 hrs") %>% column_to_rownames()
subdata <- subdata[,-c(27237:27239)]
log.data.FPKM.sample.info <- t(subdata)
for(i in 1:ncol(log.data.FPKM.sample.info)){
for(j in 1:ncol(log.data.FPKM.sample.info)){
x = log.data.FPKM.sample.info[,i]
y = log.data.FPKM.sample.info[,j]
## M-values
M=x-y ## A-values
A = (x + y)/2
df = data.frame(A, M)
cat('###',"MA plot", colnames(log.data.FPKM.sample.info)[[i]], "-", colnames(log.data.FPKM.sample.info)[[j]] ,' \n')
p <- ggplot(df, aes(x = A, y = M)) + geom_point(size = 1.5, alpha = 1/5) +
geom_hline(color = "blue3", yintercept = 0) + stat_smooth(se = FALSE, method = "loess", color = "red3") + theme_bw()
print(p)
#print(htmltools::tagList(ggplotly(p)))
cat("\n \n")
}
}MA plot A016 - A016
MA plot A016 - A142
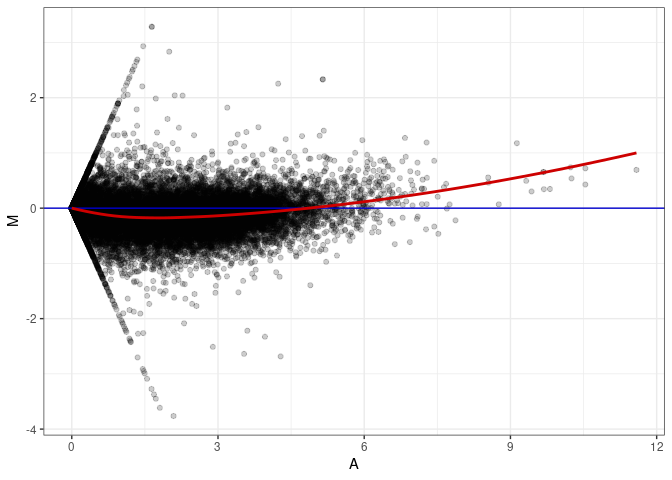
MA plot A016 - A151
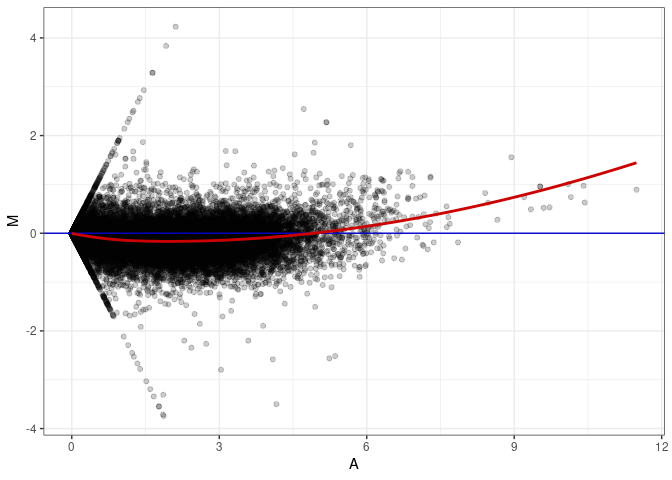
MA plot A016 - A169
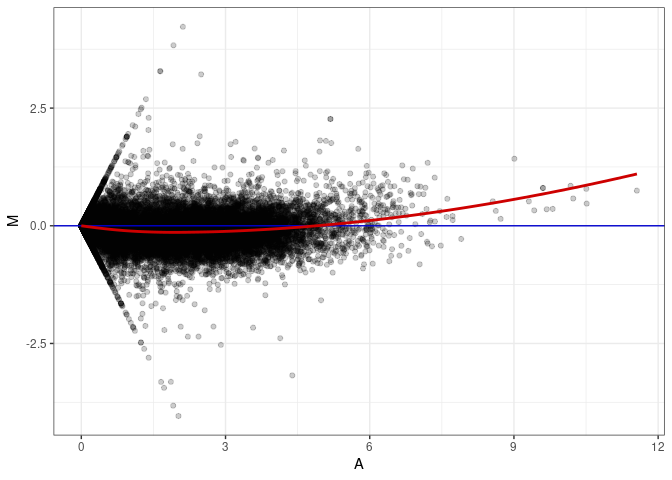
MA plot A142 - A016
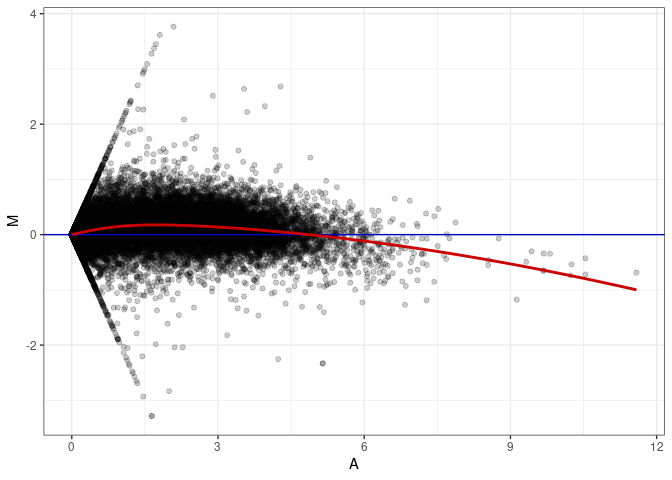
MA plot A142 - A142
MA plot A142 - A151
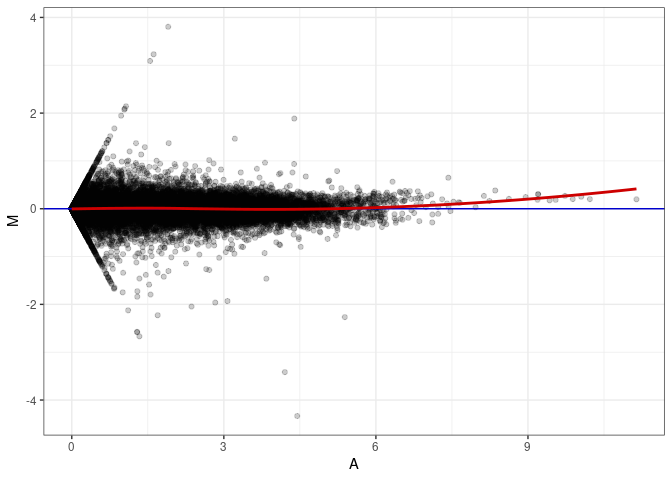
MA plot A142 - A169
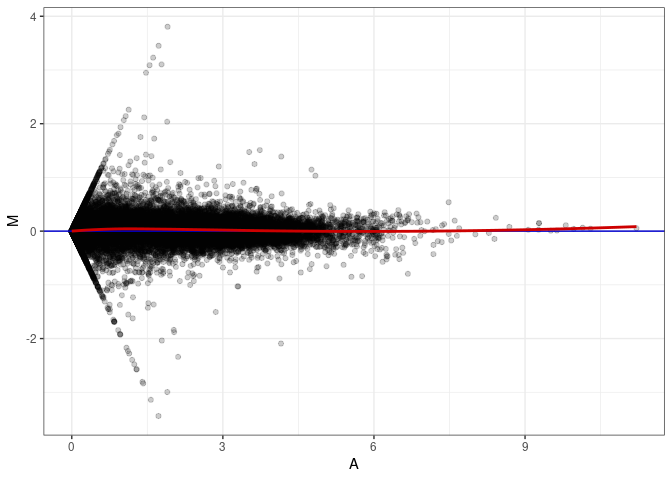
MA plot A151 - A016
MA plot A151 - A142
MA plot A151 - A151
MA plot A151 - A169
MA plot A169 - A016
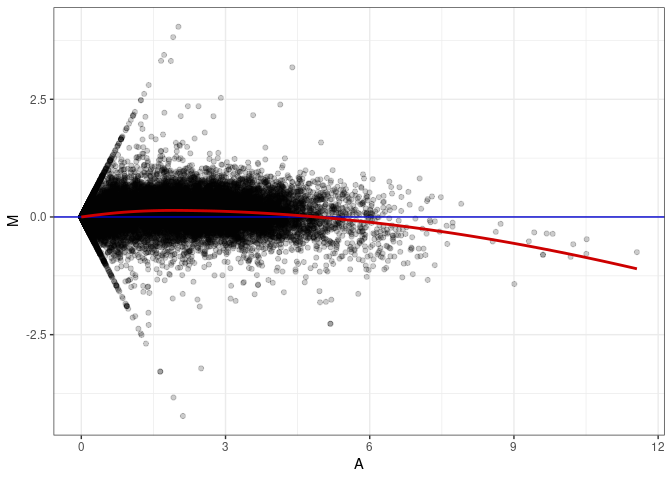
MA plot A169 - A142
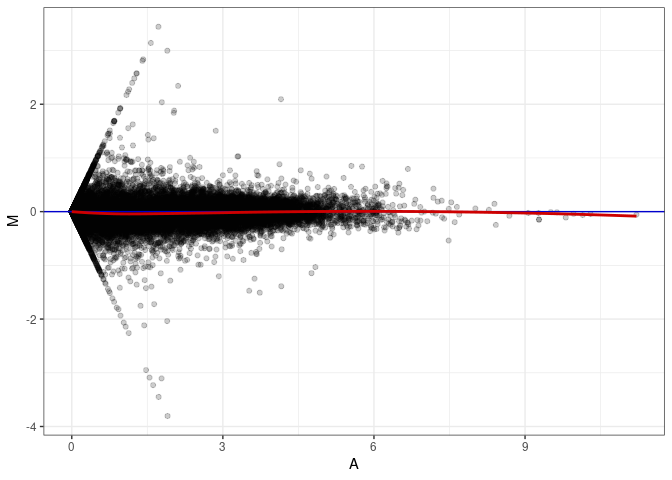
MA plot A169 - A151
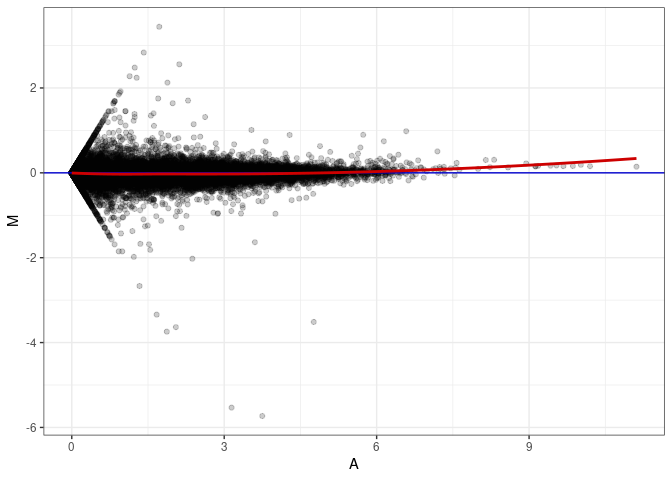
MA plot A169 - A169
MA plot for Hypothalamus control 96hrs
subdata <- log.tdata.FPKM.sample.info %>% rownames_to_column() %>% filter(Tissue =="Hypothanamus" & Treatment == "None" & Time == "96 hrs") %>% column_to_rownames()
subdata <- subdata[,-c(27237:27239)]
log.data.FPKM.sample.info <- t(subdata)
for(i in 1:ncol(log.data.FPKM.sample.info)){
for(j in 1:ncol(log.data.FPKM.sample.info)){
x = log.data.FPKM.sample.info[,i]
y = log.data.FPKM.sample.info[,j]
## M-values
M=x-y ## A-values
A = (x + y)/2
df = data.frame(A, M)
cat('###',"MA plot", colnames(log.data.FPKM.sample.info)[[i]], "-", colnames(log.data.FPKM.sample.info)[[j]] ,' \n')
p <- ggplot(df, aes(x = A, y = M)) + geom_point(size = 1.5, alpha = 1/5) +
geom_hline(color = "blue3", yintercept = 0) + stat_smooth(se = FALSE, method = "loess", color = "red3") + theme_bw()
print(p)
#print(htmltools::tagList(ggplotly(p)))
cat("\n \n")
}
}MA plot A061 - A061
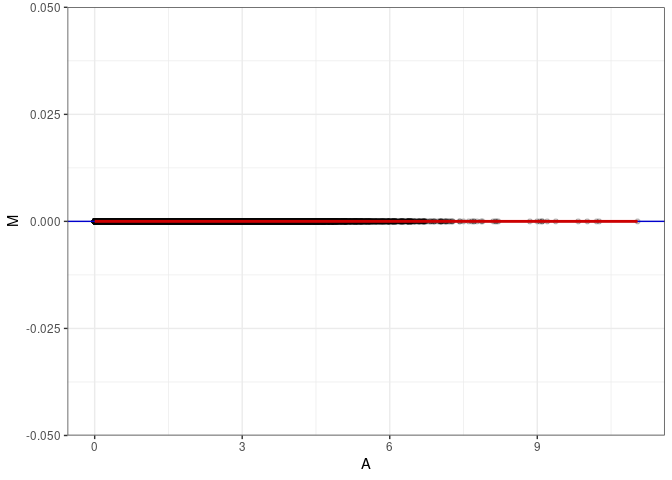
MA plot A061 - A070
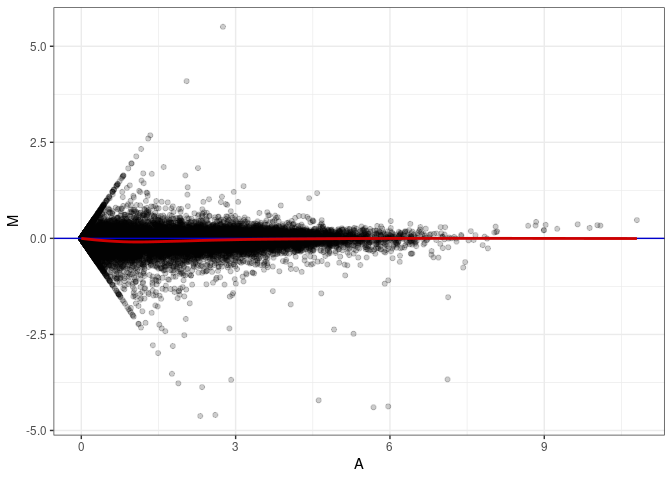
MA plot A061 - A079
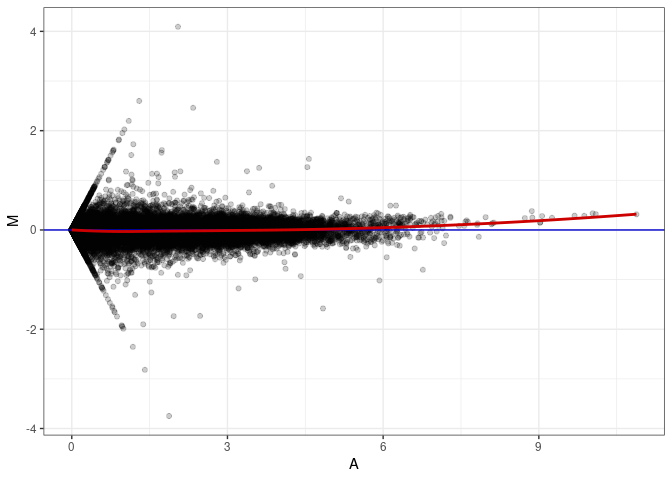
MA plot A061 - A115
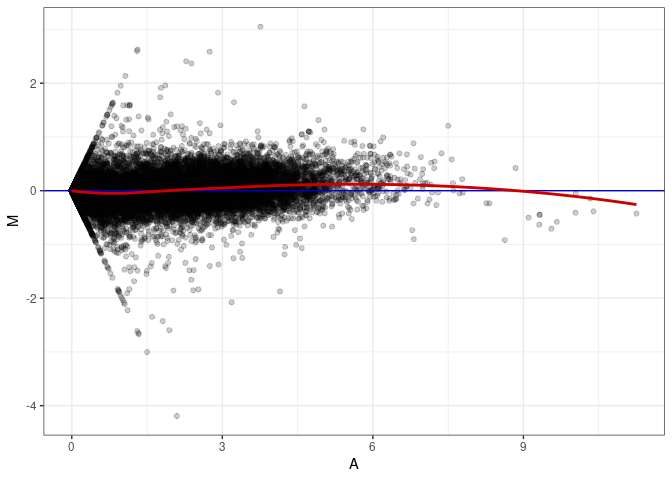
MA plot A070 - A061
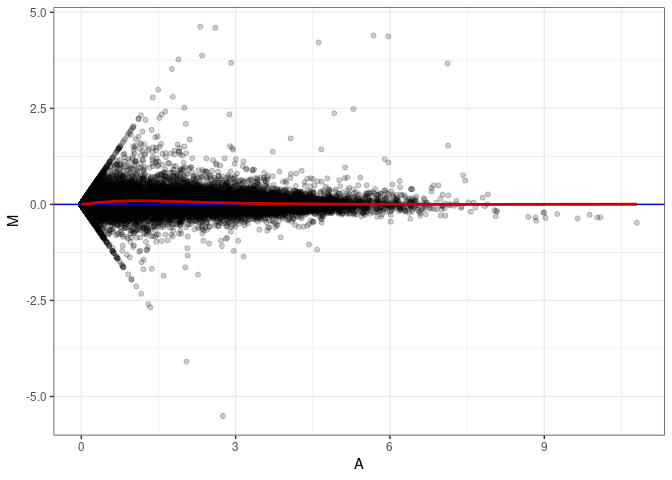
MA plot A070 - A070
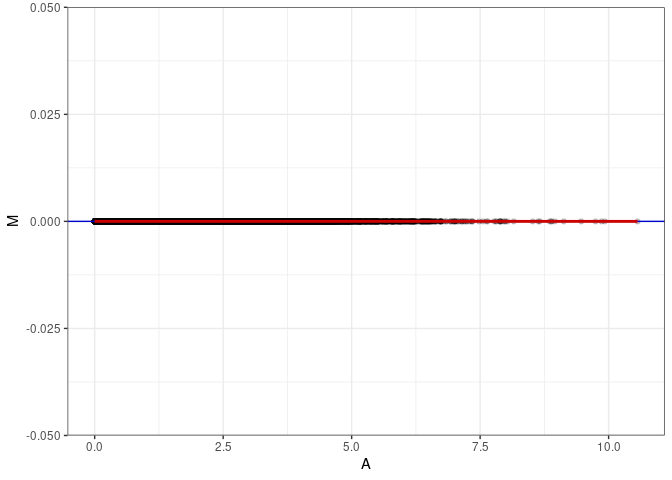
MA plot A070 - A079
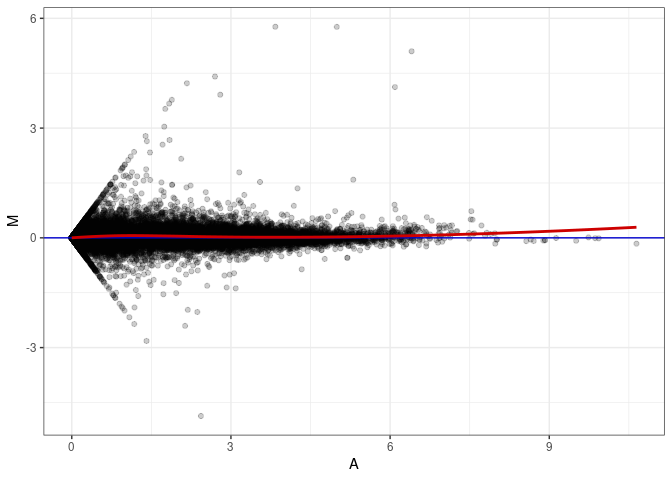
MA plot A070 - A115
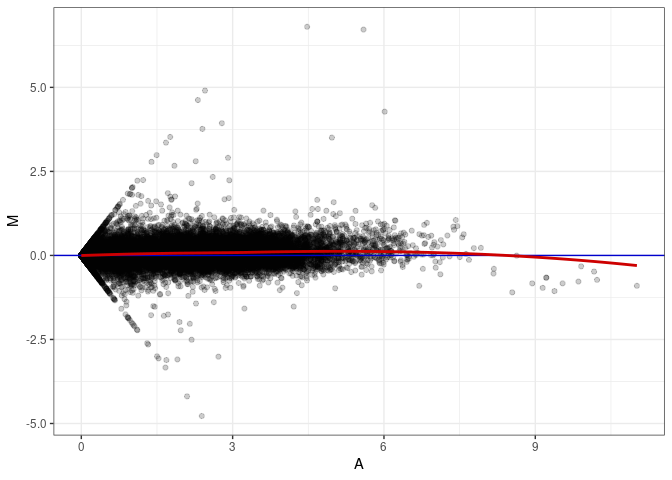
MA plot A079 - A061
MA plot A079 - A070
MA plot A079 - A079
MA plot A079 - A115
MA plot A115 - A061
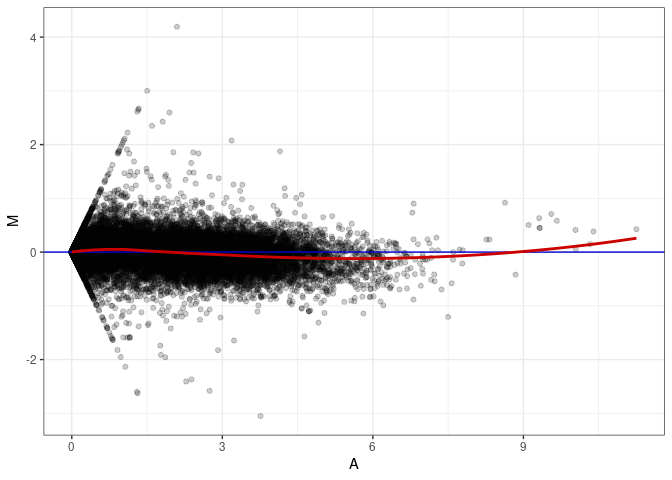
MA plot A115 - A070
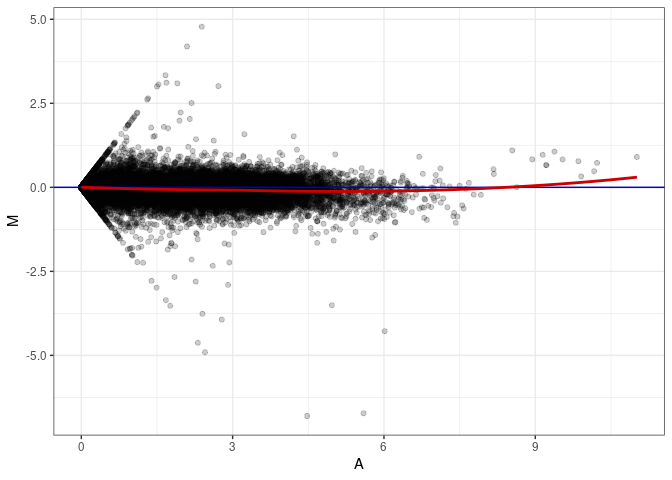
MA plot A115 - A079
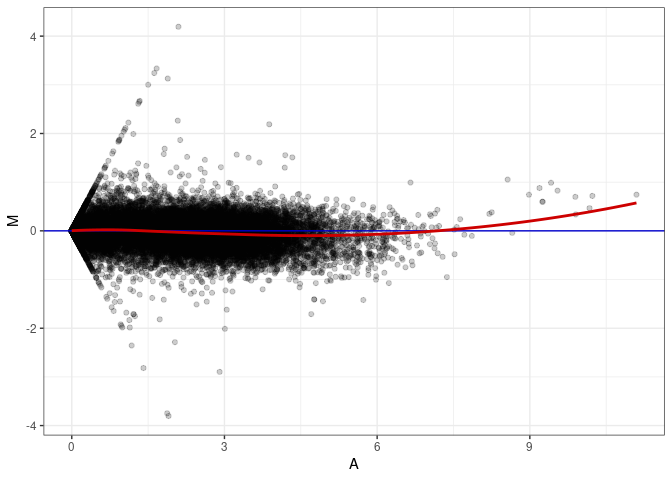
MA plot A115 - A115
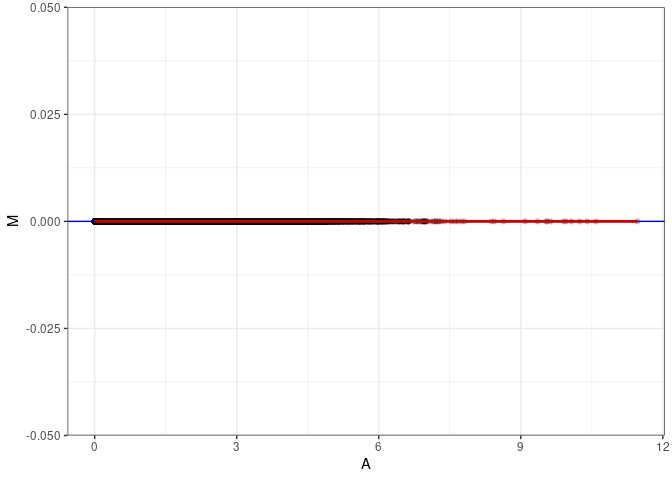
MA plot for Hippocampus 2DG 4wks
subdata <- log.tdata.FPKM.sample.info %>% rownames_to_column() %>% filter(Tissue =="Hippocampus" & Treatment == "2DG" & Time == "4 wks") %>% column_to_rownames()
subdata <- subdata[,-c(27237:27239)]
log.data.FPKM.sample.info <- t(subdata)
for(i in 1:ncol(log.data.FPKM.sample.info)){
for(j in 1:ncol(log.data.FPKM.sample.info)){
x = log.data.FPKM.sample.info[,i]
y = log.data.FPKM.sample.info[,j]
## M-values
M=x-y ## A-values
A = (x + y)/2
df = data.frame(A, M)
cat('###',"MA plot", colnames(log.data.FPKM.sample.info)[[i]], "-", colnames(log.data.FPKM.sample.info)[[j]] ,' \n')
p <- ggplot(df, aes(x = A, y = M)) + geom_point(size = 1.5, alpha = 1/5) +
geom_hline(color = "blue3", yintercept = 0) + stat_smooth(se = FALSE, method = "loess", color = "red3") + theme_bw()
print(p)
#print(htmltools::tagList(ggplotly(p)))
cat("\n \n")
}
}MA plot A026 - A026
MA plot A026 - A035
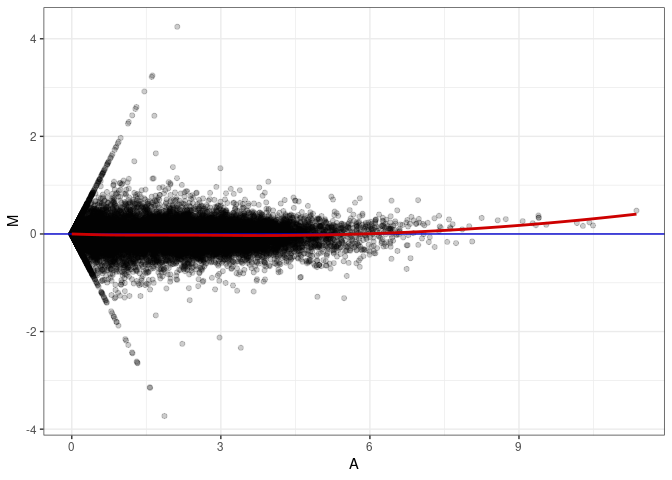
MA plot A026 - A098
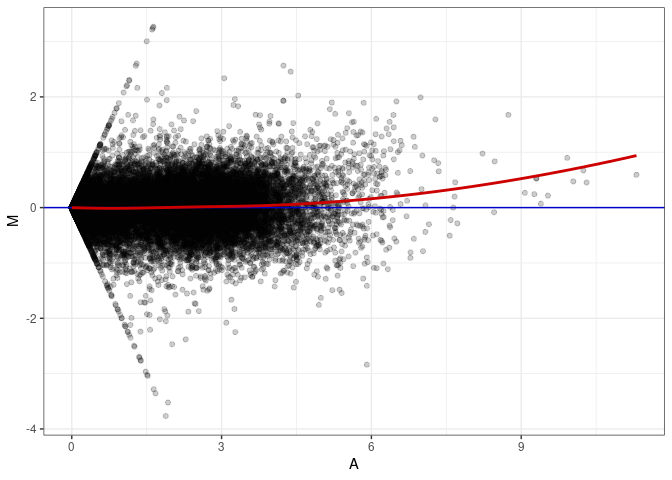
MA plot A026 - A179
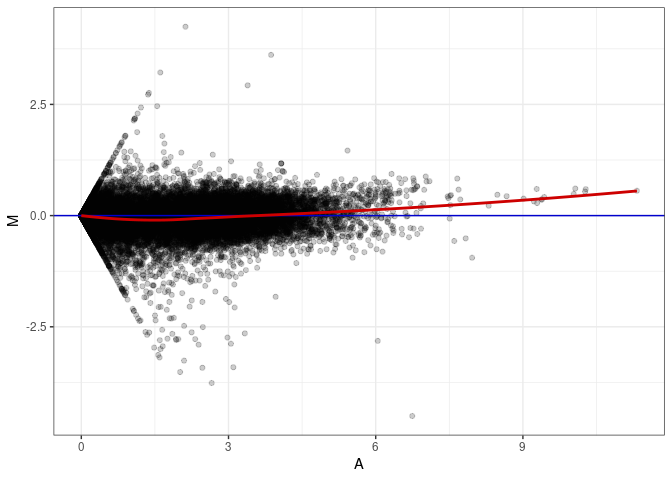
MA plot A035 - A026
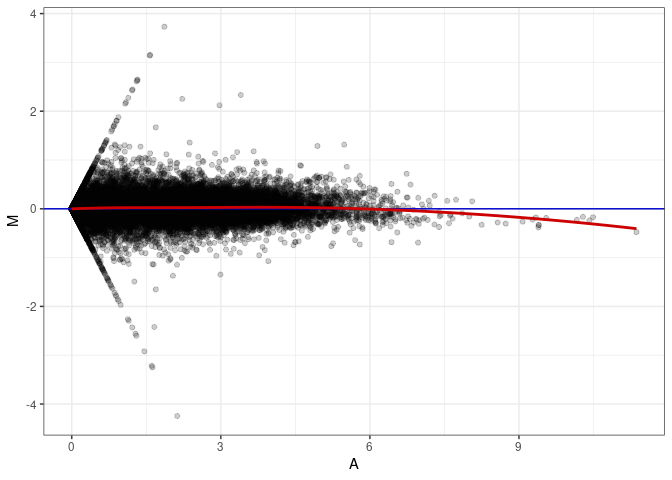
MA plot A035 - A035
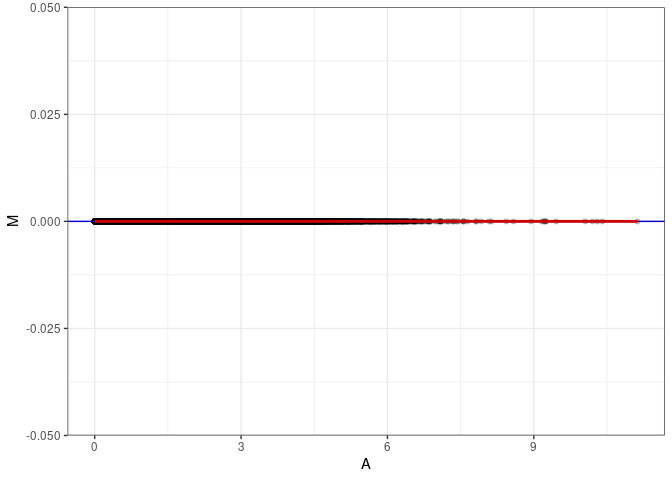
MA plot A035 - A098
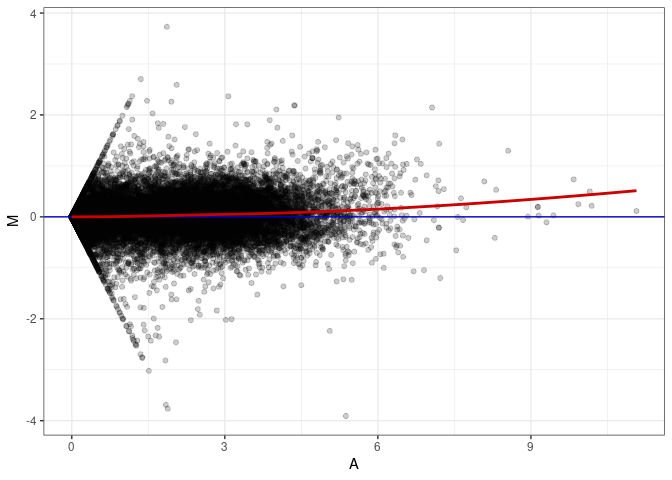
MA plot A035 - A179
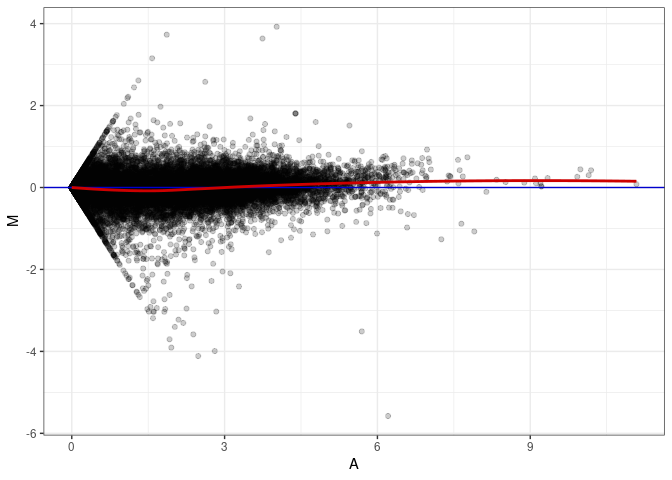
MA plot A098 - A026
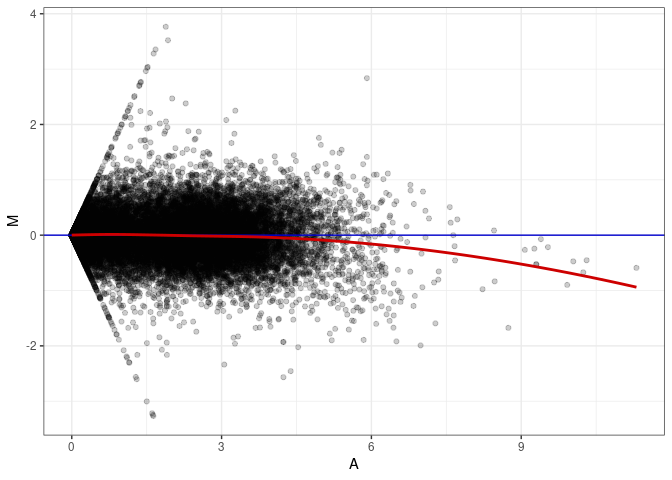
MA plot A098 - A035
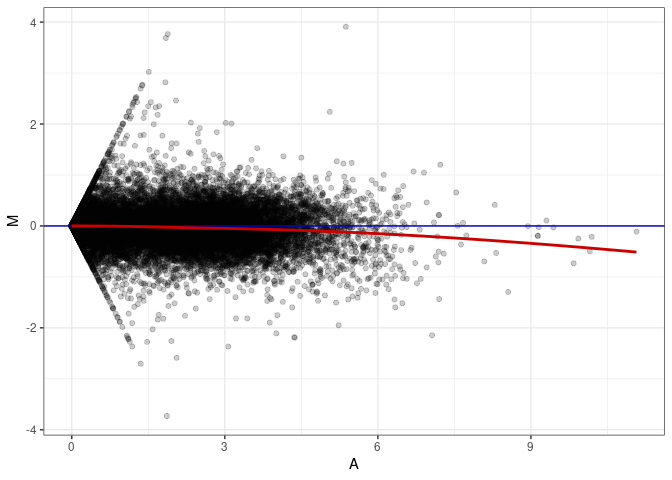
MA plot A098 - A098
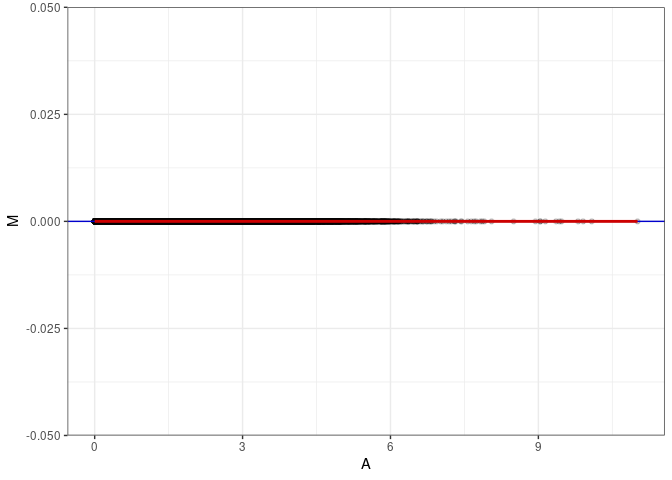
MA plot A098 - A179
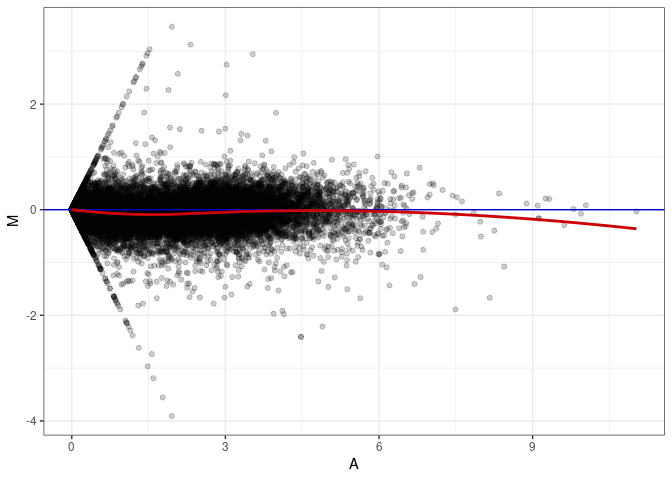
MA plot A179 - A026
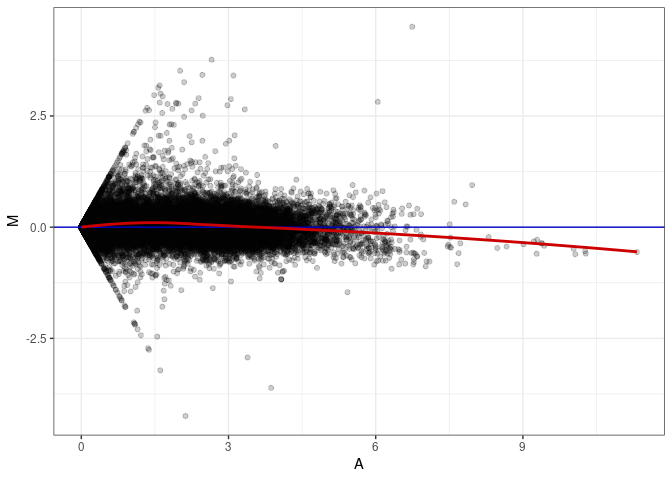
MA plot A179 - A035
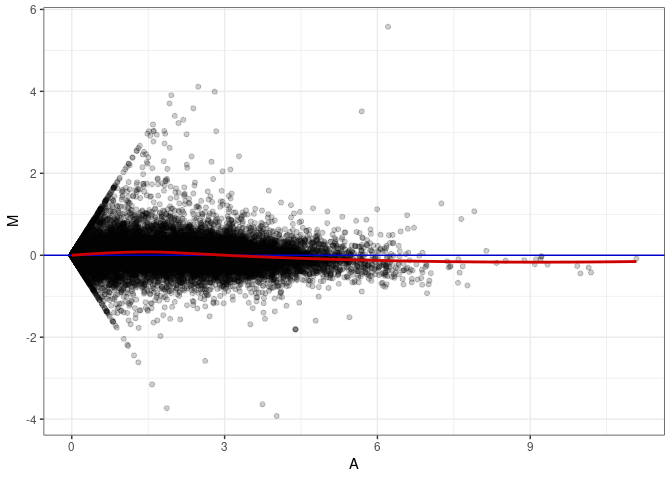
MA plot A179 - A098
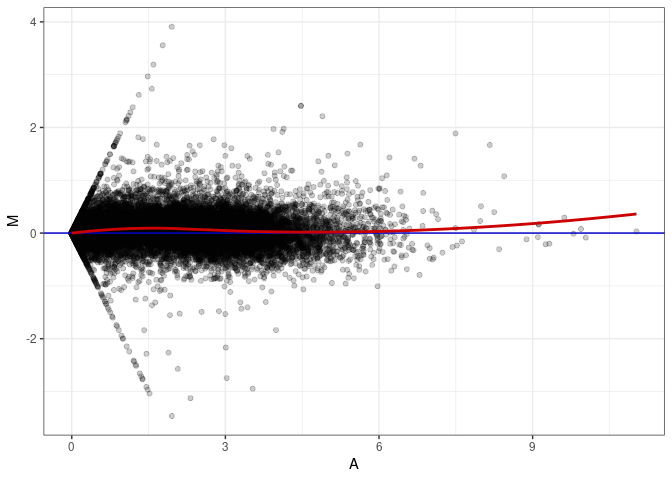
MA plot A179 - A179
MA plot for Hippocampus control 4wks
subdata <- log.tdata.FPKM.sample.info %>% rownames_to_column() %>% filter(Tissue =="Hippocampus" & Treatment == "None" & Time == "4 wks") %>% column_to_rownames()
subdata <- subdata[,-c(27237:27239)]
log.data.FPKM.sample.info <- t(subdata)
for(i in 1:ncol(log.data.FPKM.sample.info)){
for(j in 1:ncol(log.data.FPKM.sample.info)){
x = log.data.FPKM.sample.info[,i]
y = log.data.FPKM.sample.info[,j]
## M-values
M=x-y ## A-values
A = (x + y)/2
df = data.frame(A, M)
cat('###',"MA plot", colnames(log.data.FPKM.sample.info)[[i]], "-", colnames(log.data.FPKM.sample.info)[[j]] ,' \n')
p <- ggplot(df, aes(x = A, y = M)) + geom_point(size = 1.5, alpha = 1/5) +
geom_hline(color = "blue3", yintercept = 0) + stat_smooth(se = FALSE, method = "loess", color = "red3") + theme_bw()
print(p)
#print(htmltools::tagList(ggplotly(p)))
cat("\n \n")
}
}MA plot A008 - A008
MA plot A008 - A044
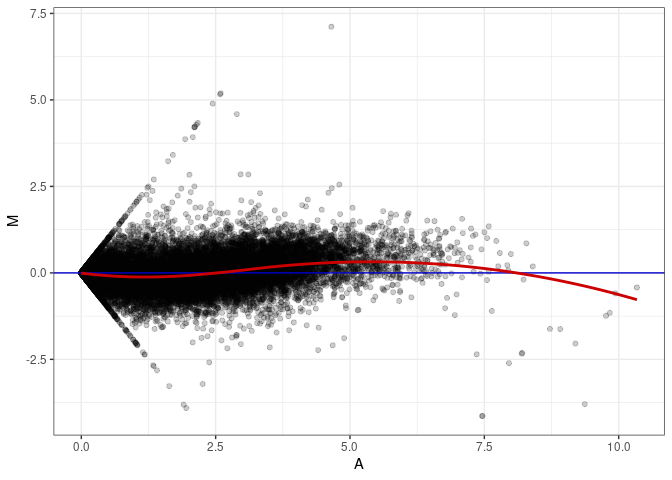
MA plot A008 - A053
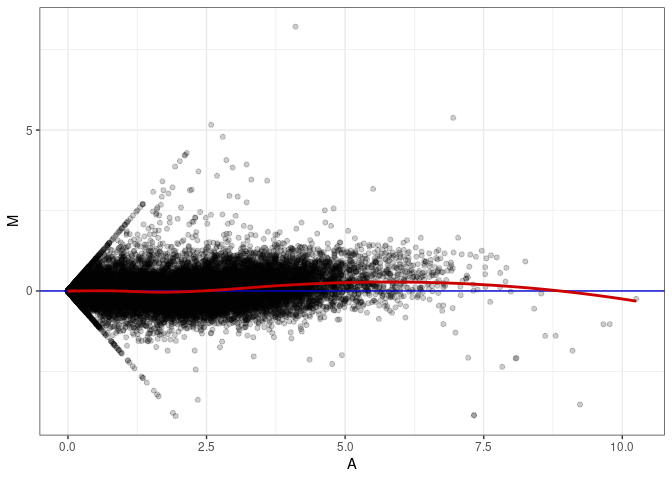
MA plot A008 - A089
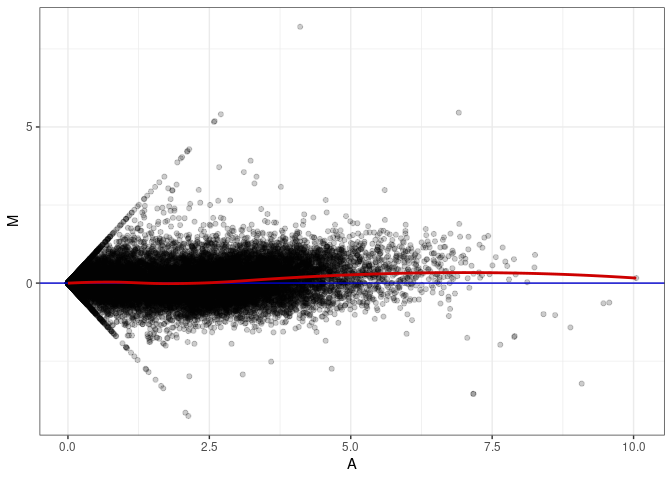
MA plot A044 - A008
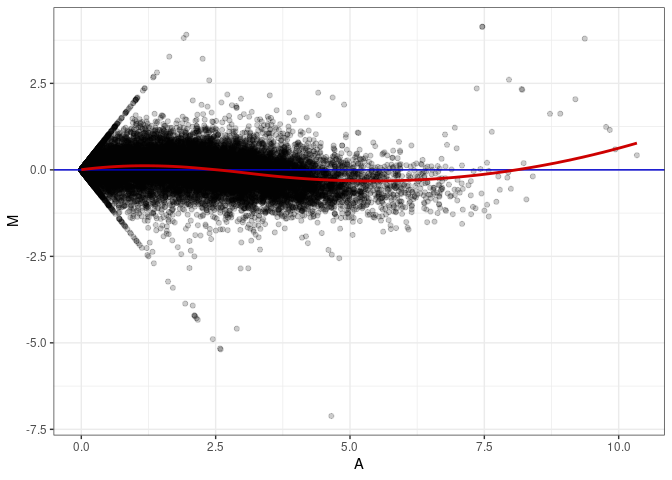
MA plot A044 - A044
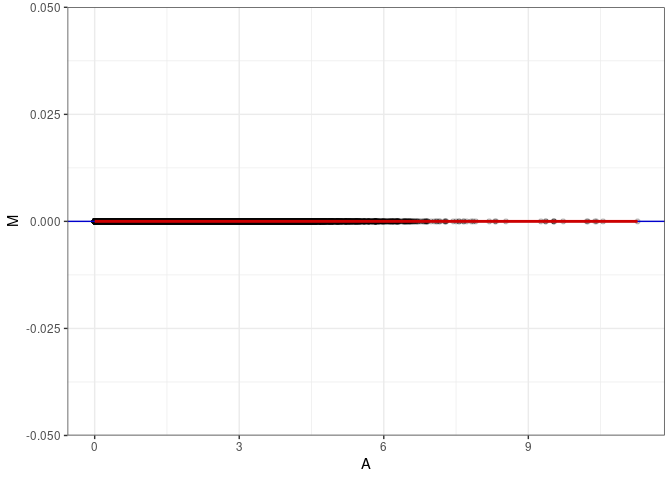
MA plot A044 - A053
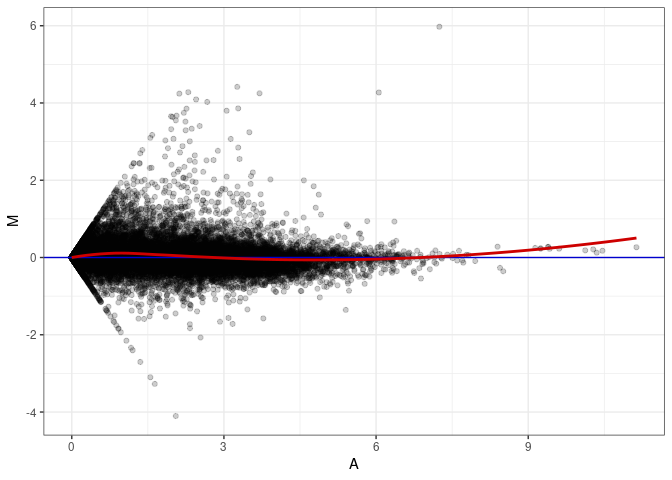
MA plot A044 - A089
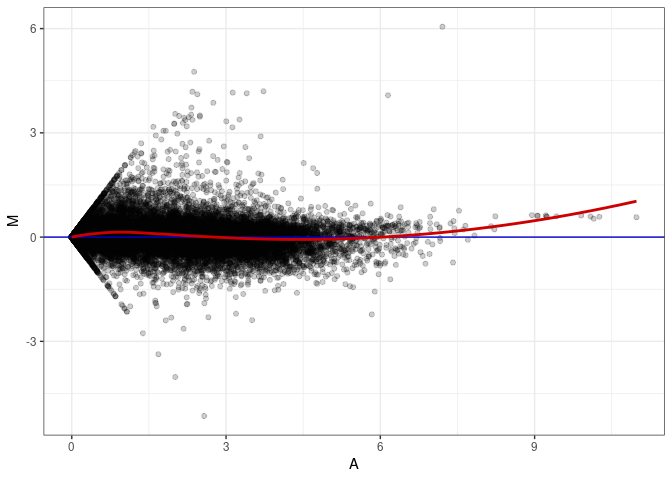
MA plot A053 - A008
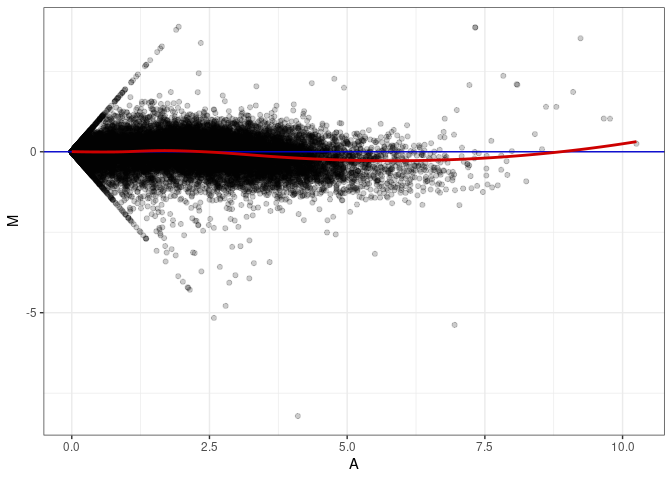
MA plot A053 - A044
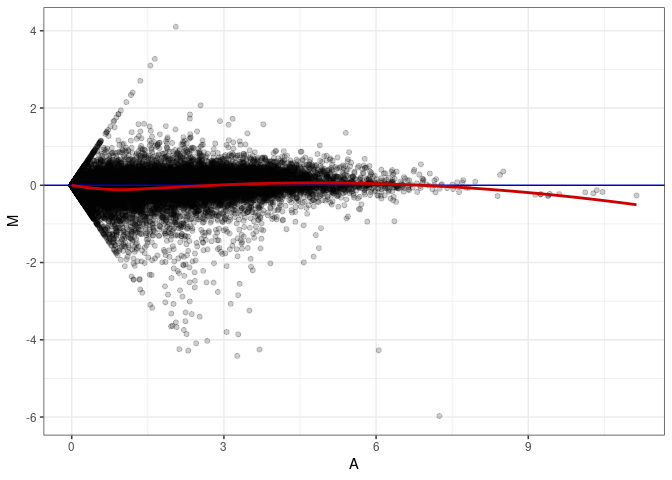
MA plot A053 - A053
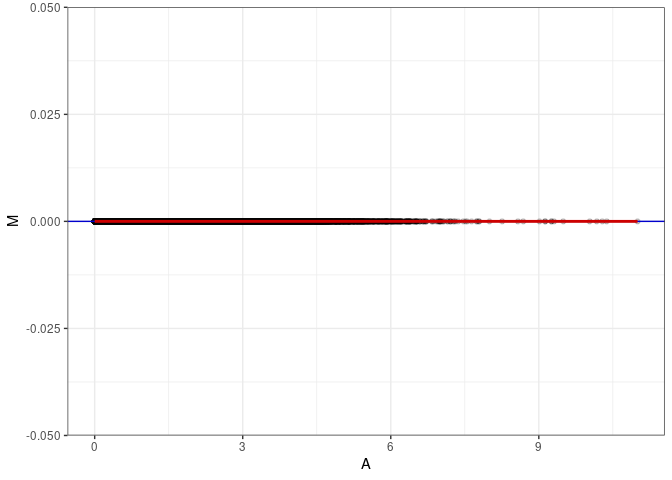
MA plot A053 - A089
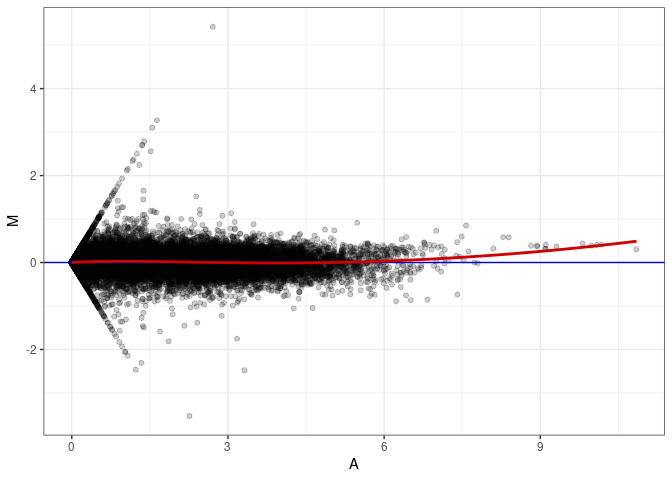
MA plot A089 - A008
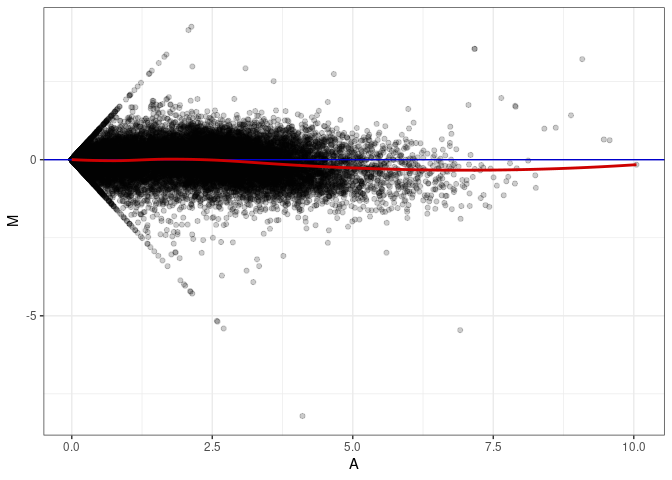
MA plot A089 - A044
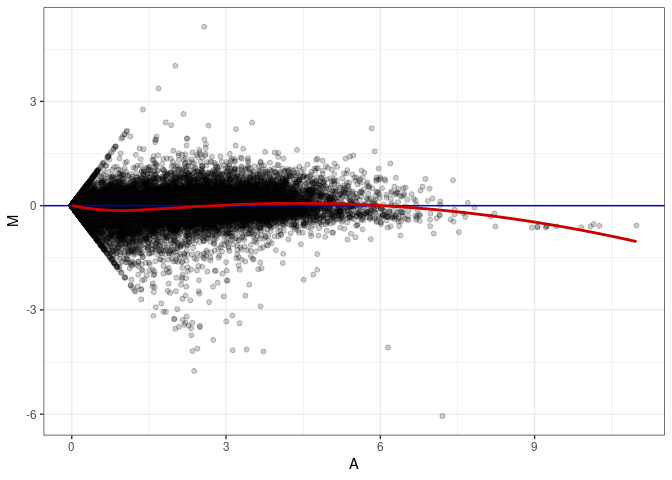
MA plot A089 - A053
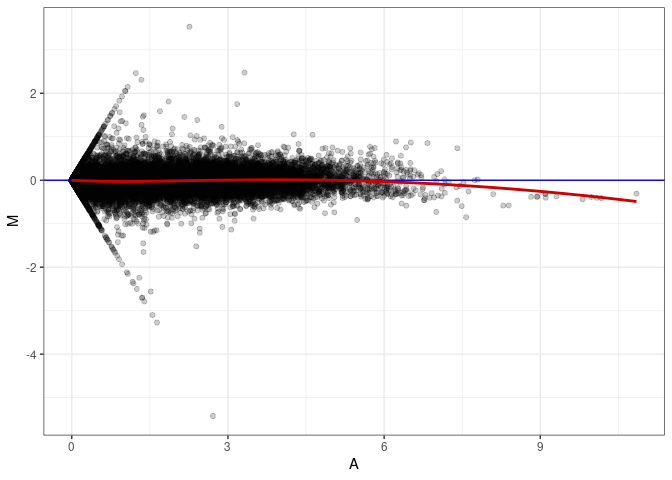
MA plot A089 - A089
MA plot for Hippocampus 2DG 96hrs
subdata <- log.tdata.FPKM.sample.info %>% rownames_to_column() %>% filter(Tissue =="Hippocampus" & Treatment == "2DG" & Time == "96 hrs") %>% column_to_rownames()
subdata <- subdata[,-c(27237:27239)]
log.data.FPKM.sample.info <- t(subdata)
for(i in 1:ncol(log.data.FPKM.sample.info)){
for(j in 1:ncol(log.data.FPKM.sample.info)){
x = log.data.FPKM.sample.info[,i]
y = log.data.FPKM.sample.info[,j]
## M-values
M=x-y ## A-values
A = (x + y)/2
df = data.frame(A, M)
cat('###',"MA plot", colnames(log.data.FPKM.sample.info)[[i]], "-", colnames(log.data.FPKM.sample.info)[[j]] ,' \n')
p <- ggplot(df, aes(x = A, y = M)) + geom_point(size = 1.5, alpha = 1/5) +
geom_hline(color = "blue3", yintercept = 0) + stat_smooth(se = FALSE, method = "loess", color = "red3") + theme_bw()
print(p)
#print(htmltools::tagList(ggplotly(p)))
cat("\n \n")
}
}MA plot A017 - A017
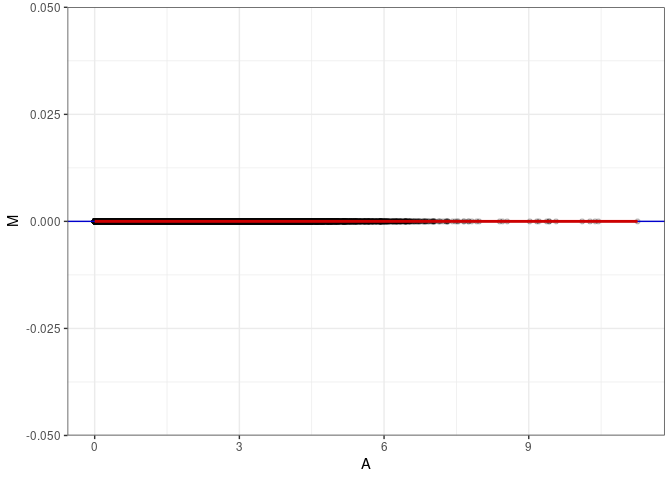
MA plot A017 - A143
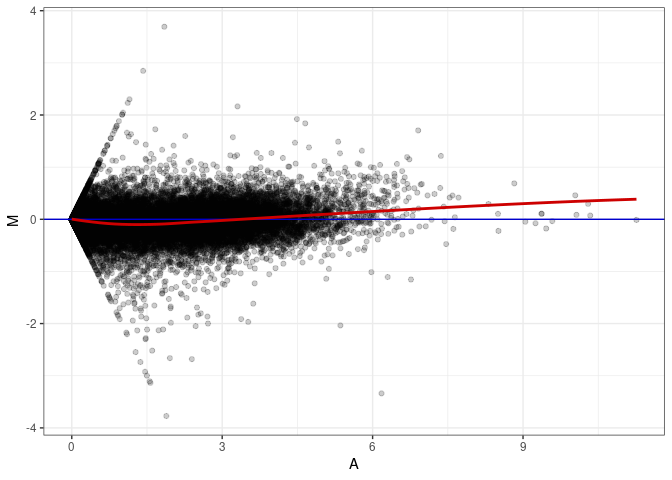
MA plot A017 - A152
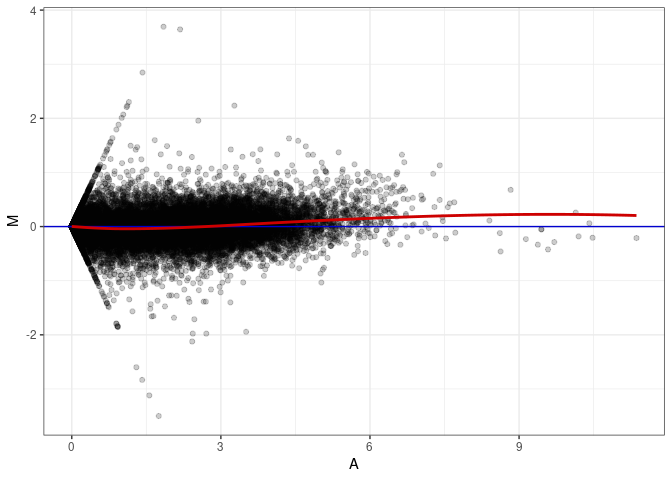
MA plot A017 - A170
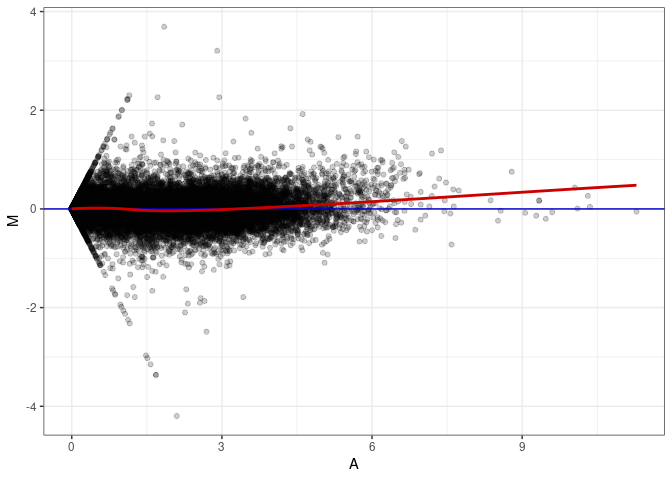
MA plot A143 - A017
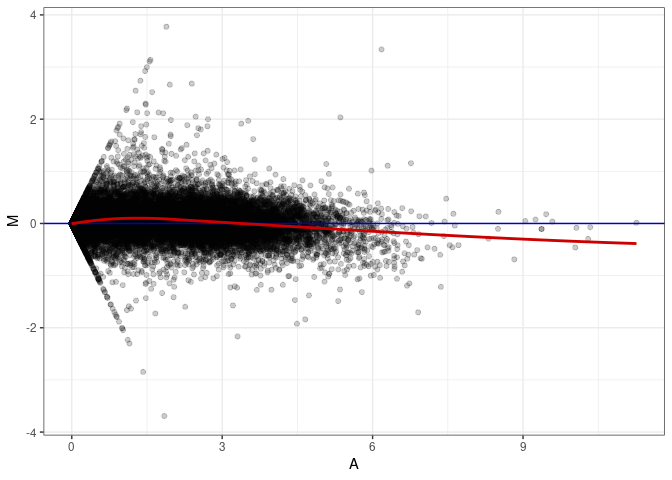
MA plot A143 - A143
MA plot A143 - A152
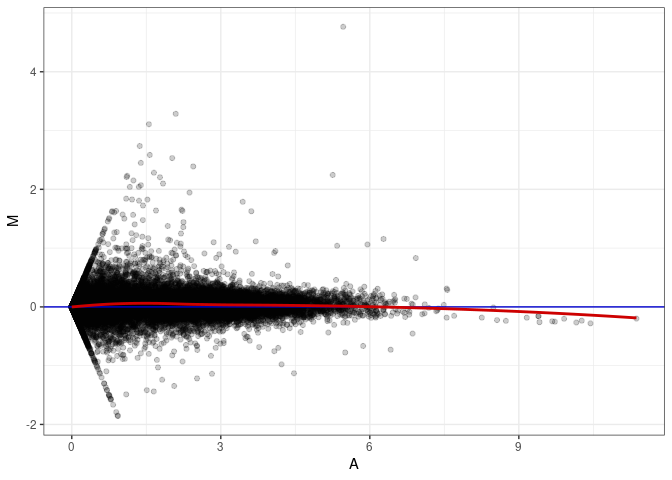
MA plot A143 - A170
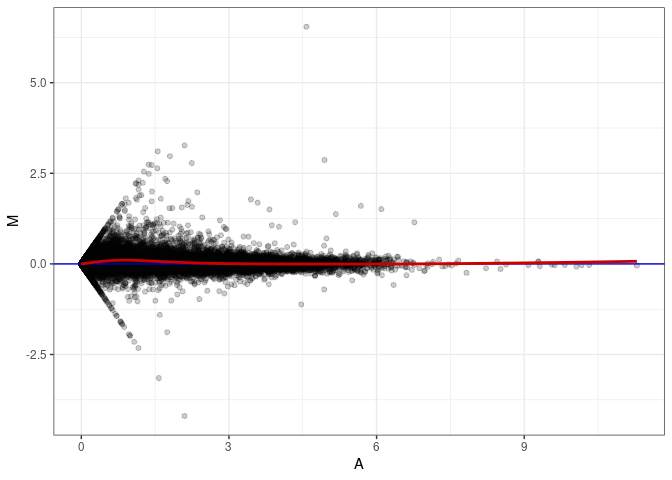
MA plot A152 - A017
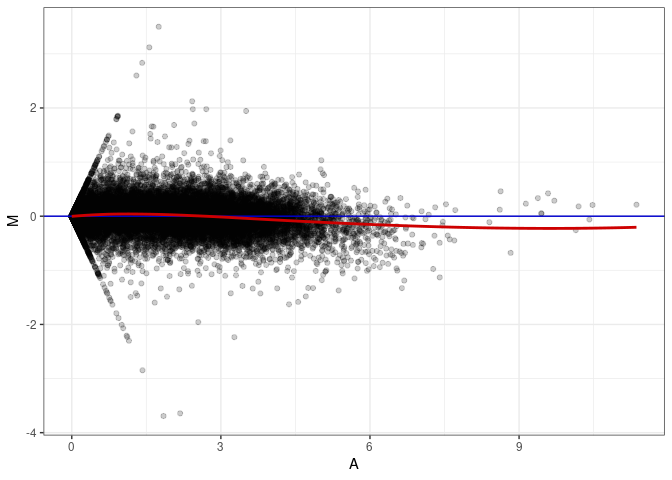
MA plot A152 - A143
MA plot A152 - A152
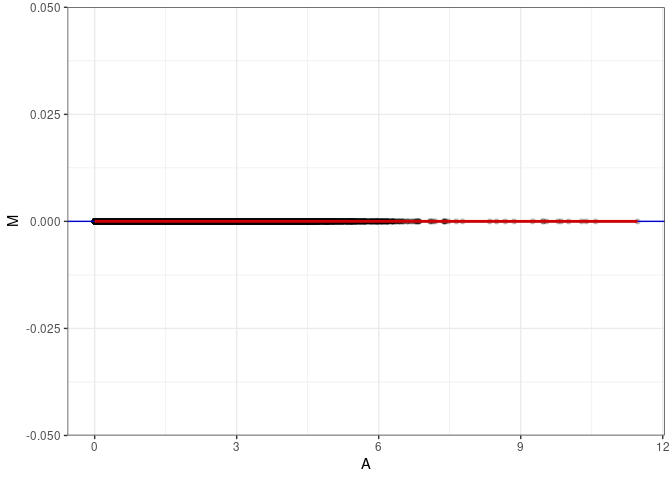
MA plot A152 - A170
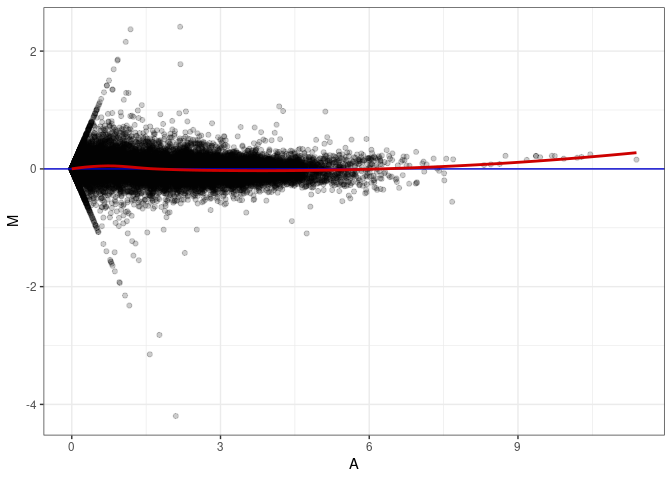
MA plot A170 - A017
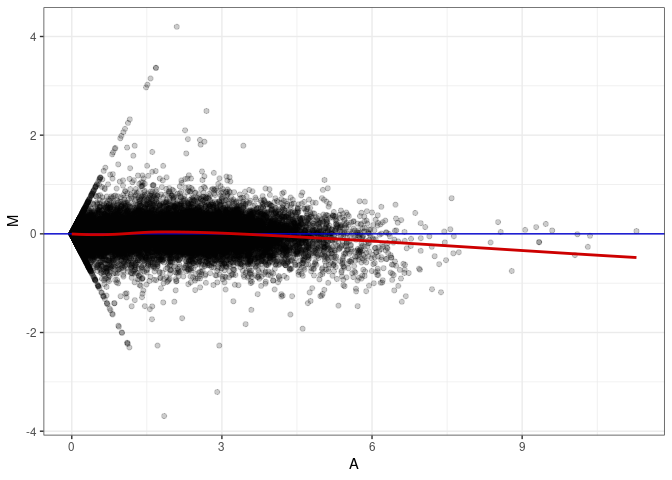
MA plot A170 - A143
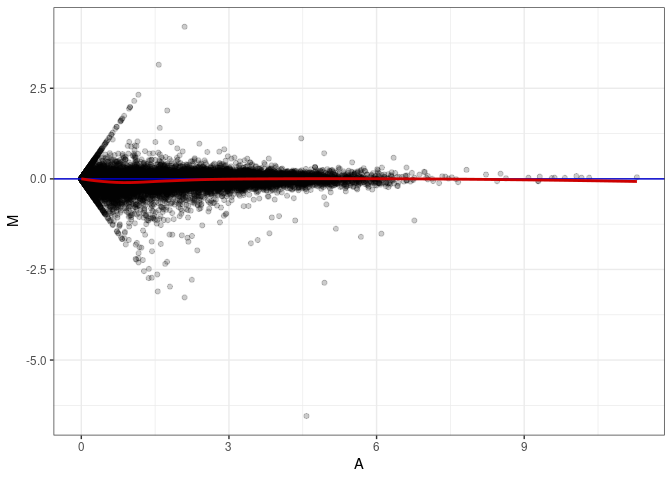
MA plot A170 - A152
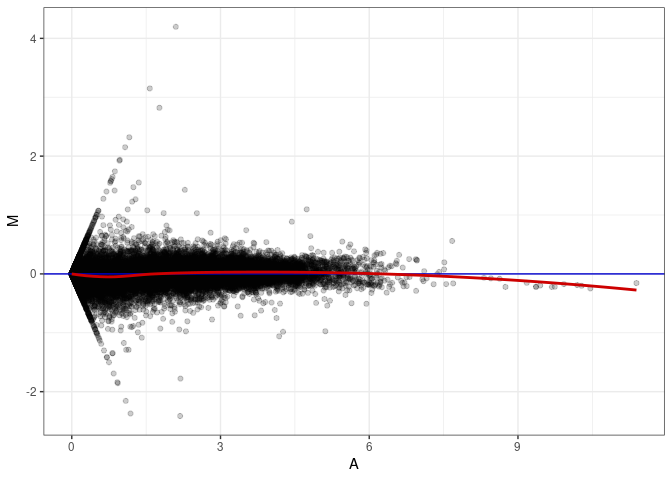
MA plot A170 - A170
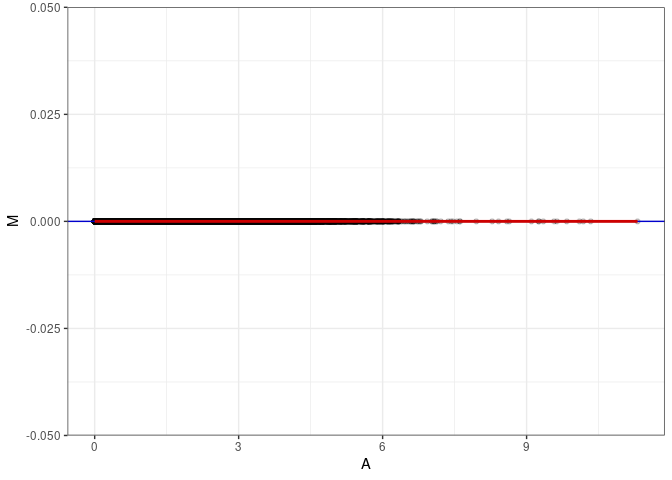
MA plot for Hippocampus control 96hrs
subdata <- log.tdata.FPKM.sample.info %>% rownames_to_column() %>% filter(Tissue =="Hippocampus" & Treatment == "None" & Time == "96 hrs") %>% column_to_rownames()
subdata <- subdata[,-c(27237:27239)]
log.data.FPKM.sample.info <- t(subdata)
for(i in 1:ncol(log.data.FPKM.sample.info)){
for(j in 1:ncol(log.data.FPKM.sample.info)){
x = log.data.FPKM.sample.info[,i]
y = log.data.FPKM.sample.info[,j]
## M-values
M=x-y ## A-values
A = (x + y)/2
df = data.frame(A, M)
cat('###',"MA plot", colnames(log.data.FPKM.sample.info)[[i]], "-", colnames(log.data.FPKM.sample.info)[[j]] ,' \n')
p <- ggplot(df, aes(x = A, y = M)) + geom_point(size = 1.5, alpha = 1/5) +
geom_hline(color = "blue3", yintercept = 0) + stat_smooth(se = FALSE, method = "loess", color = "red3") + theme_bw()
print(p)
#print(htmltools::tagList(ggplotly(p)))
cat("\n \n")
}
}MA plot A062 - A062
MA plot A062 - A071
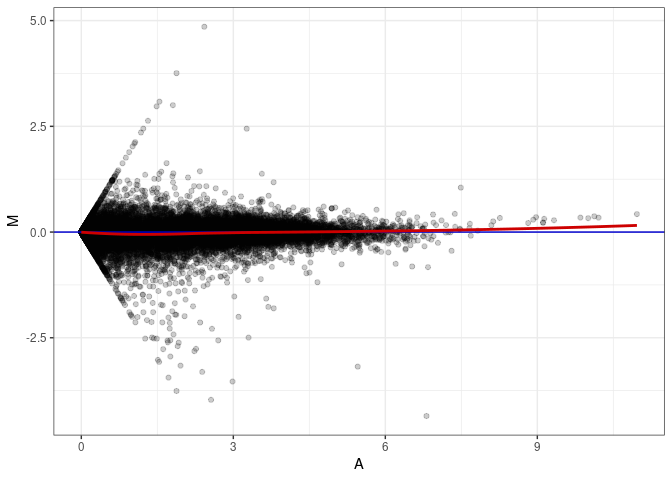
MA plot A062 - A080
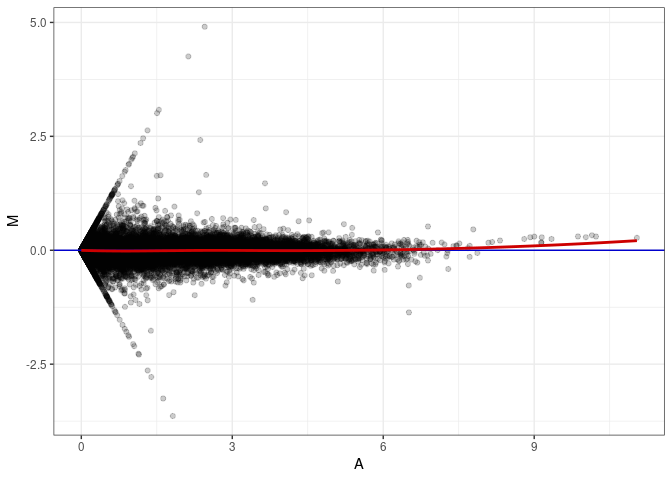
MA plot A062 - A116
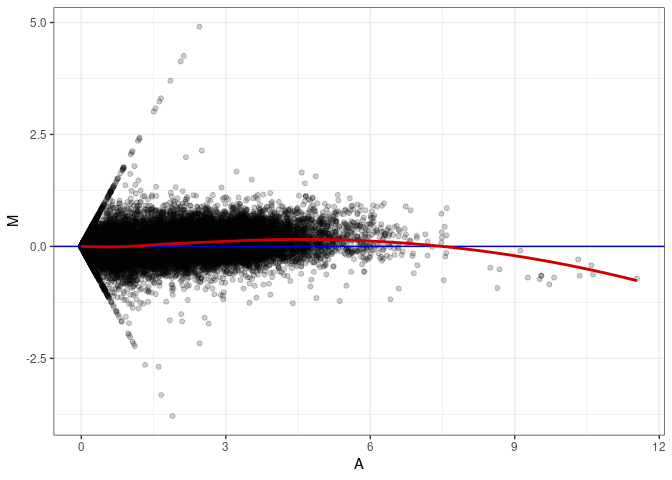
MA plot A071 - A062
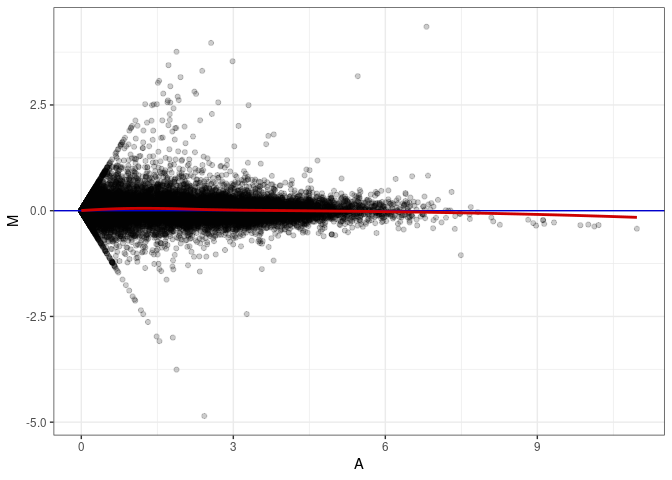
MA plot A071 - A071
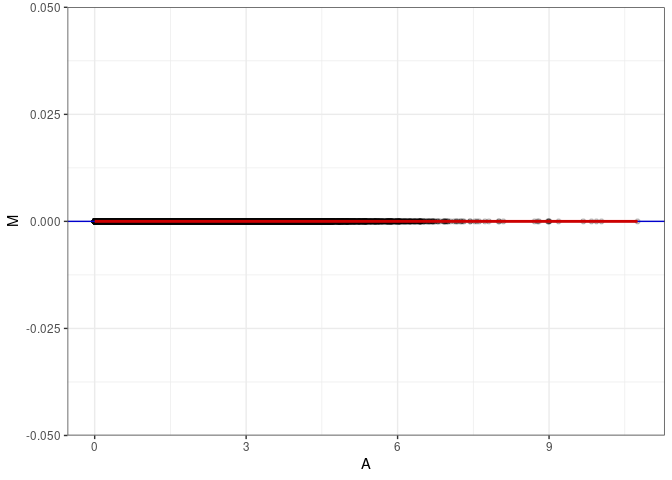
MA plot A071 - A080
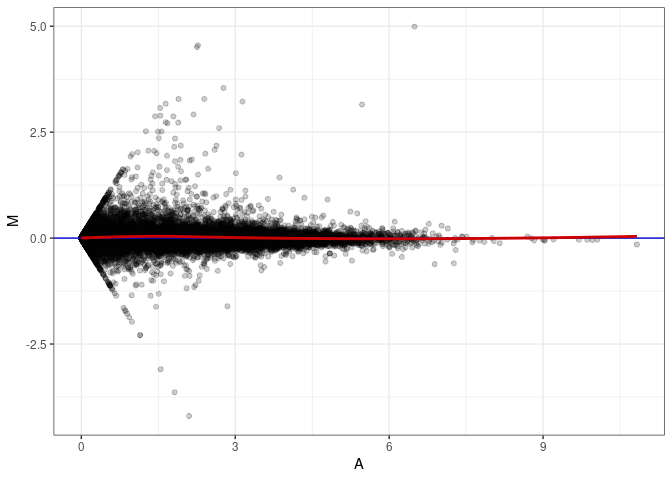
MA plot A071 - A116
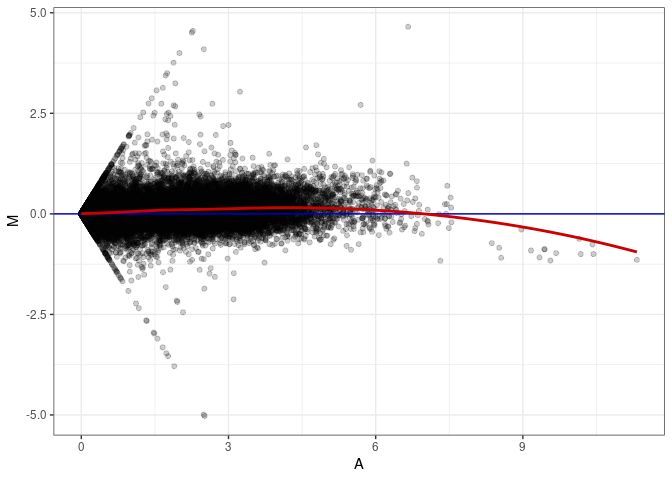
MA plot A080 - A062
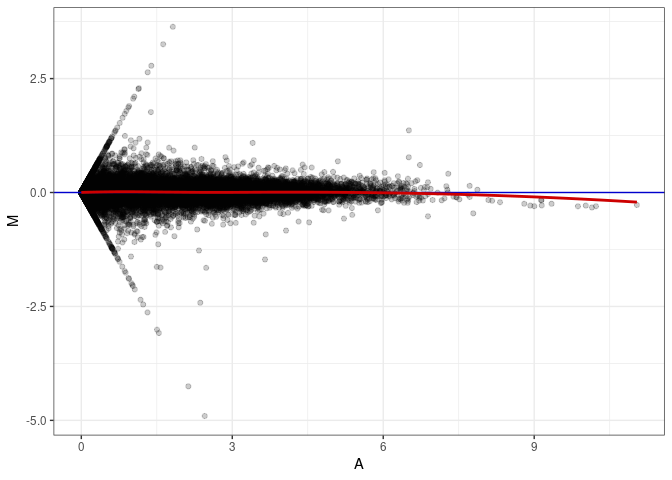
MA plot A080 - A071
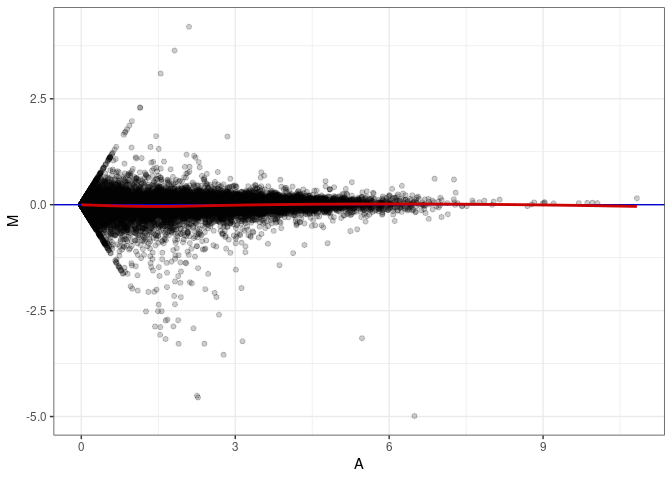
MA plot A080 - A080
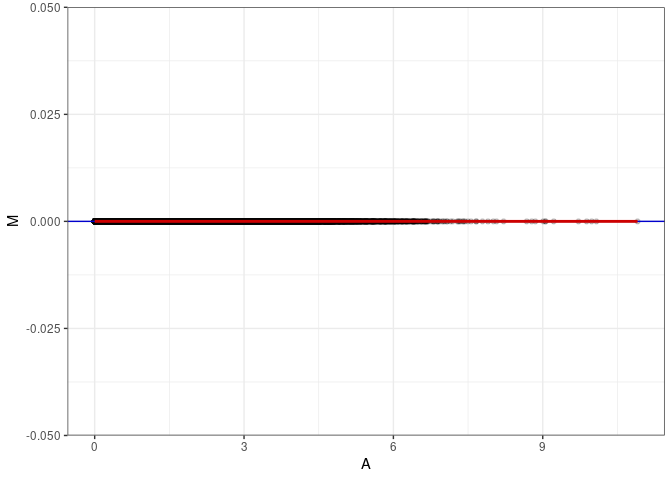
MA plot A080 - A116
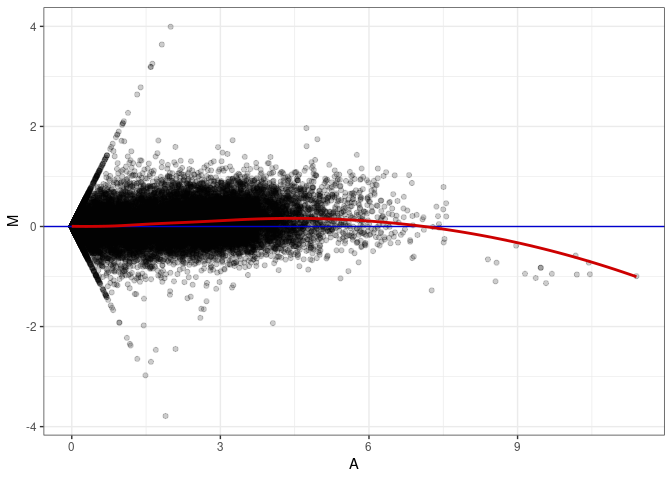
MA plot A116 - A062
MA plot A116 - A071
MA plot A116 - A080
MA plot A116 - A116
MA plot for Liver 2DG 4wks
subdata <- log.tdata.FPKM.sample.info %>% rownames_to_column() %>% filter(Tissue =="Liver" & Treatment == "2DG" & Time == "4 wks") %>% column_to_rownames()
subdata <- subdata[,-c(27237:27239)]
log.data.FPKM.sample.info <- t(subdata)
for(i in 1:ncol(log.data.FPKM.sample.info)){
for(j in 1:ncol(log.data.FPKM.sample.info)){
x = log.data.FPKM.sample.info[,i]
y = log.data.FPKM.sample.info[,j]
## M-values
M=x-y ## A-values
A = (x + y)/2
df = data.frame(A, M)
cat('###',"MA plot", colnames(log.data.FPKM.sample.info)[[i]], "-", colnames(log.data.FPKM.sample.info)[[j]] ,' \n')
p <- ggplot(df, aes(x = A, y = M)) + geom_point(size = 1.5, alpha = 1/5) +
geom_hline(color = "blue3", yintercept = 0) + stat_smooth(se = FALSE, method = "loess", color = "red3") + theme_bw()
print(p)
#print(htmltools::tagList(ggplotly(p)))
cat("\n \n")
}
}MA plot A021 - A021
MA plot A021 - A030
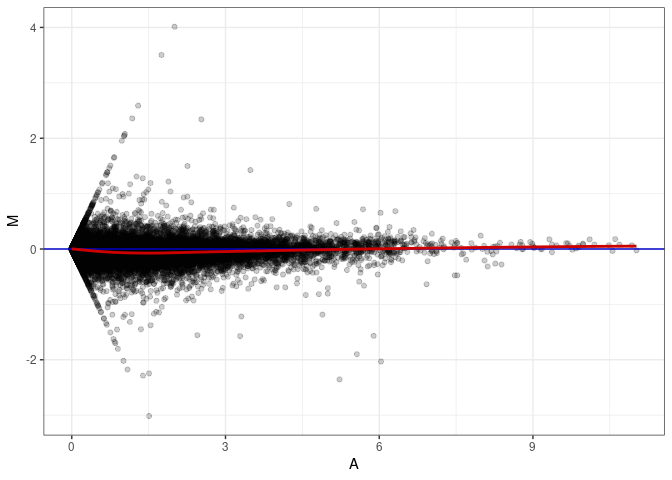
MA plot A021 - A093
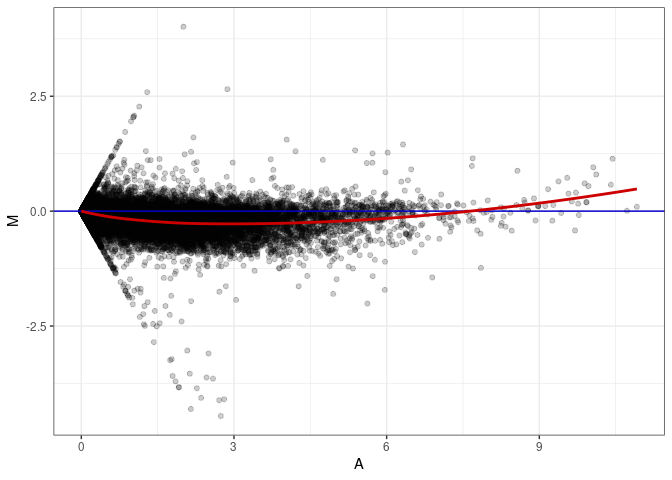
MA plot A021 - A174
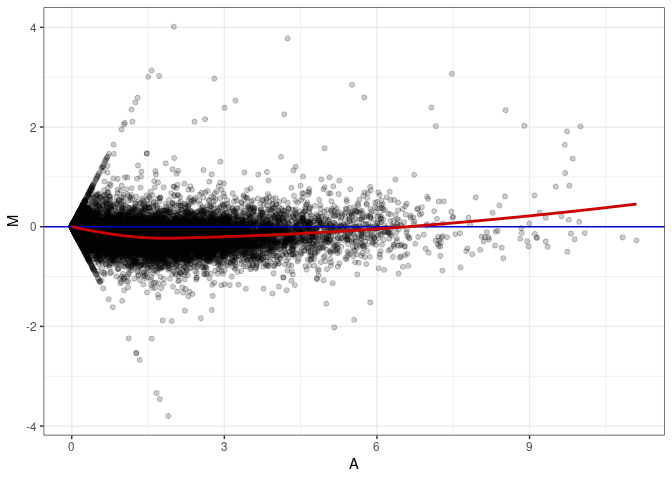
MA plot A030 - A021
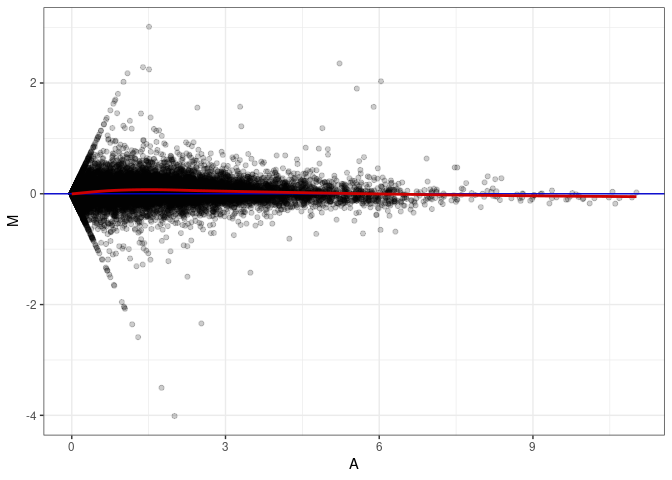
MA plot A030 - A030
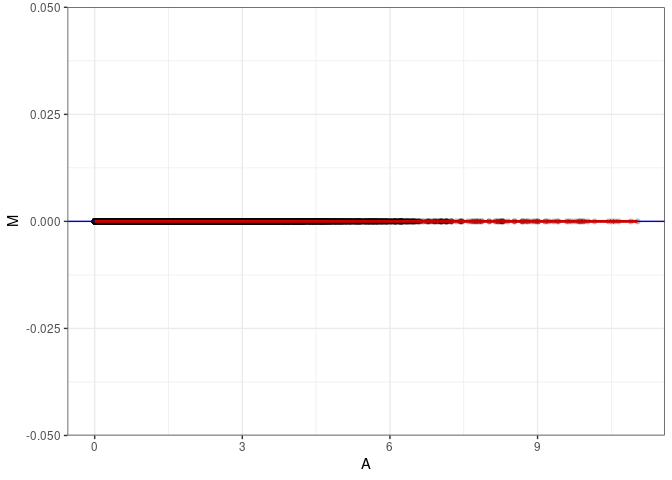
MA plot A030 - A093
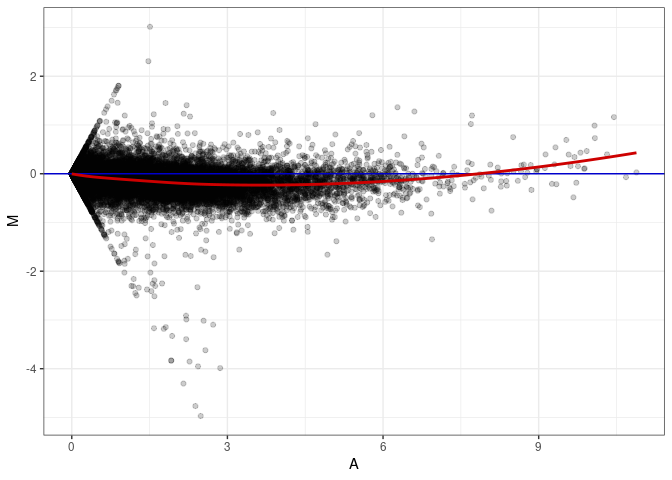
MA plot A030 - A174
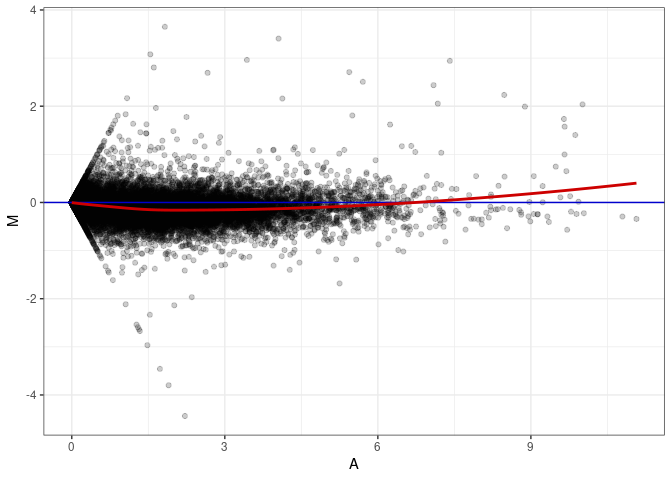
MA plot A093 - A021
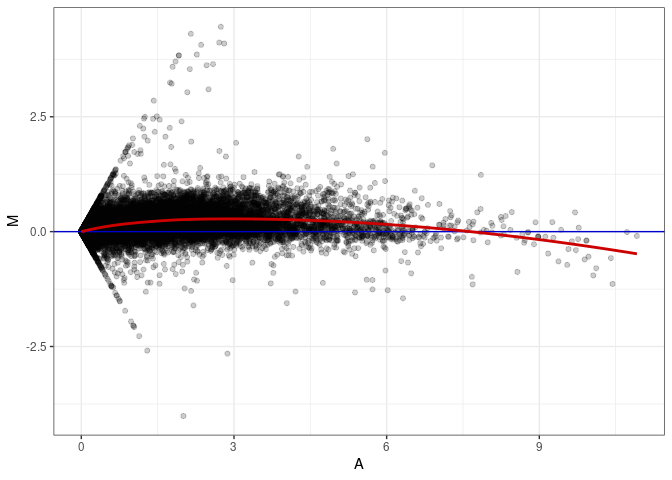
MA plot A093 - A030
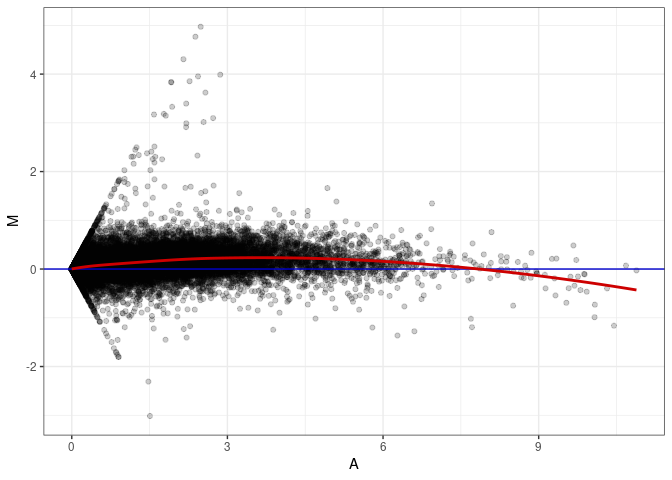
MA plot A093 - A093
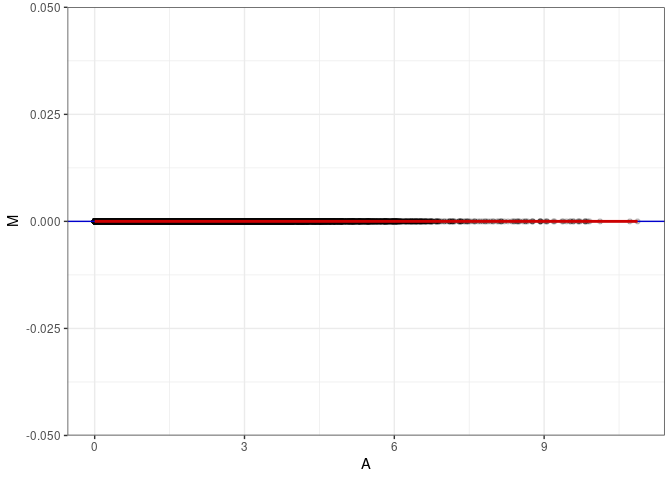
MA plot A093 - A174
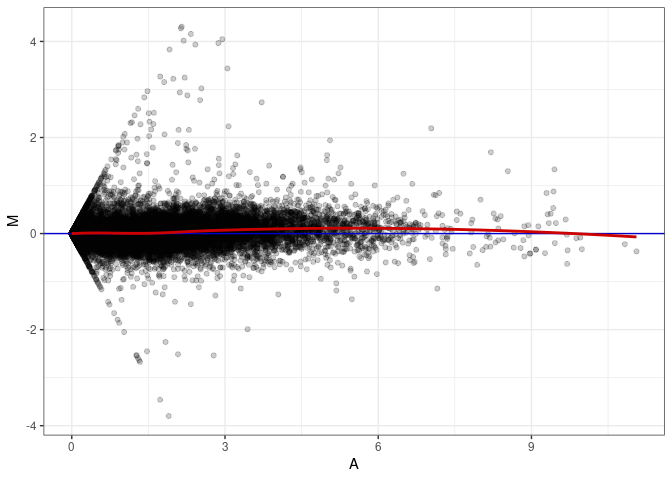
MA plot A174 - A021
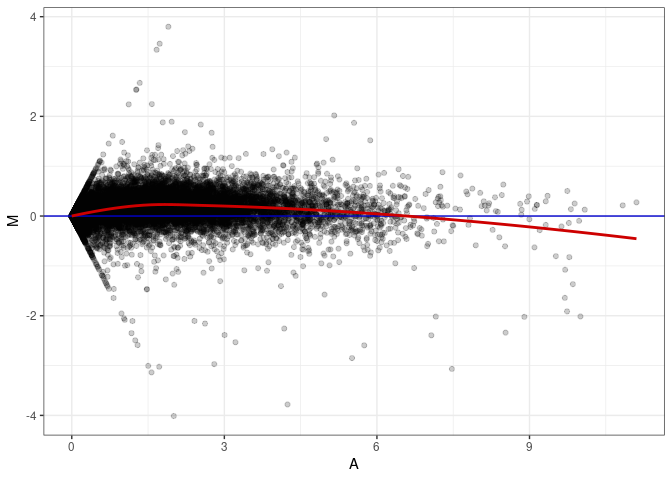
MA plot A174 - A030
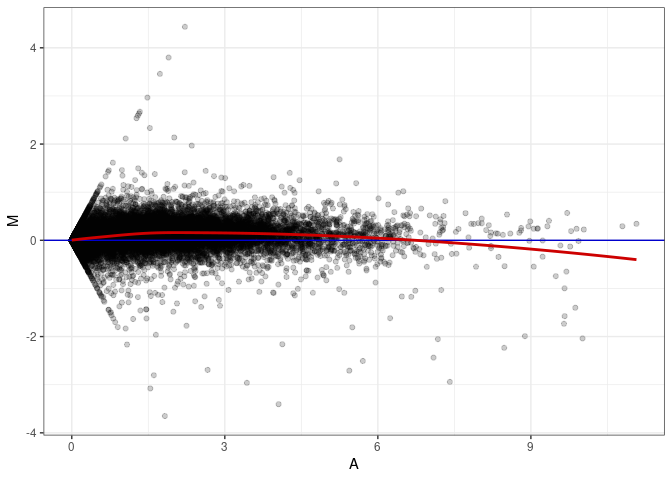
MA plot A174 - A093
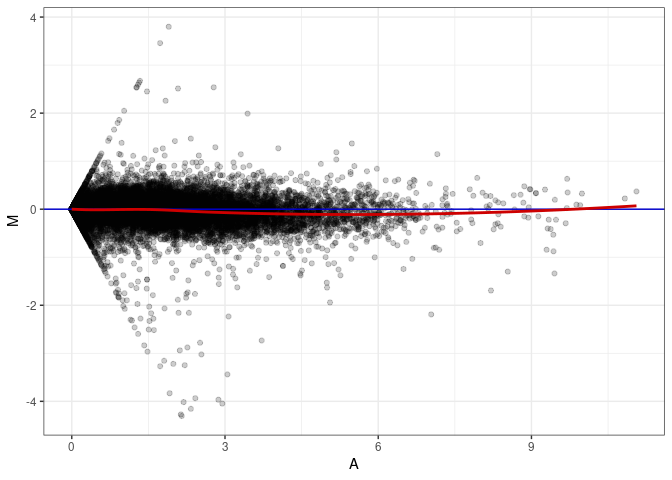
MA plot A174 - A174
MA plot for Liver control 4wks
subdata <- log.tdata.FPKM.sample.info %>% rownames_to_column() %>% filter(Tissue =="Liver" & Treatment == "None" & Time == "4 wks") %>% column_to_rownames()
subdata <- subdata[,-c(27237:27239)]
log.data.FPKM.sample.info <- t(subdata)
for(i in 1:ncol(log.data.FPKM.sample.info)){
for(j in 1:ncol(log.data.FPKM.sample.info)){
x = log.data.FPKM.sample.info[,i]
y = log.data.FPKM.sample.info[,j]
## M-values
M=x-y ## A-values
A = (x + y)/2
df = data.frame(A, M)
cat('###',"MA plot", colnames(log.data.FPKM.sample.info)[[i]], "-", colnames(log.data.FPKM.sample.info)[[j]] ,' \n')
p <- ggplot(df, aes(x = A, y = M)) + geom_point(size = 1.5, alpha = 1/5) +
geom_hline(color = "blue3", yintercept = 0) + stat_smooth(se = FALSE, method = "loess", color = "red3") + theme_bw()
print(p)
#print(htmltools::tagList(ggplotly(p)))
cat("\n \n")
}
}MA plot A003 - A003
MA plot A003 - A039
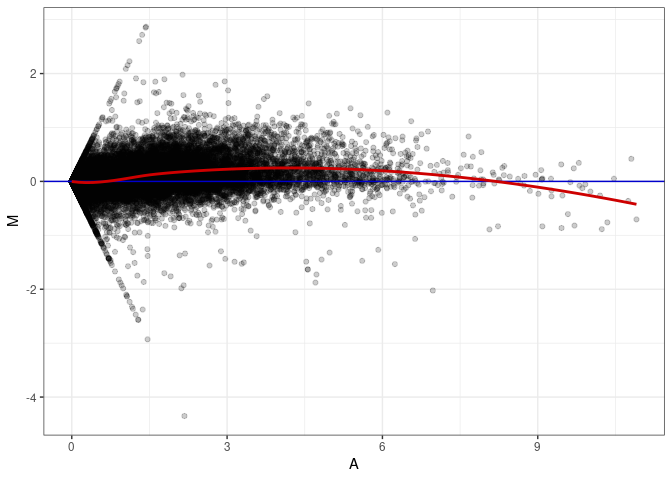
MA plot A003 - A048
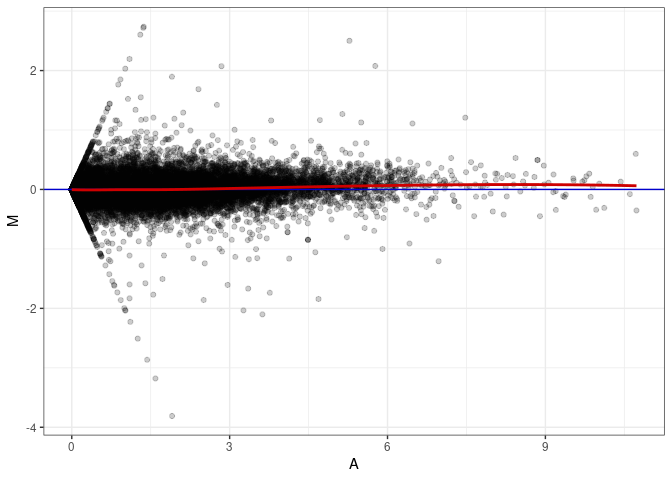
MA plot A003 - A084
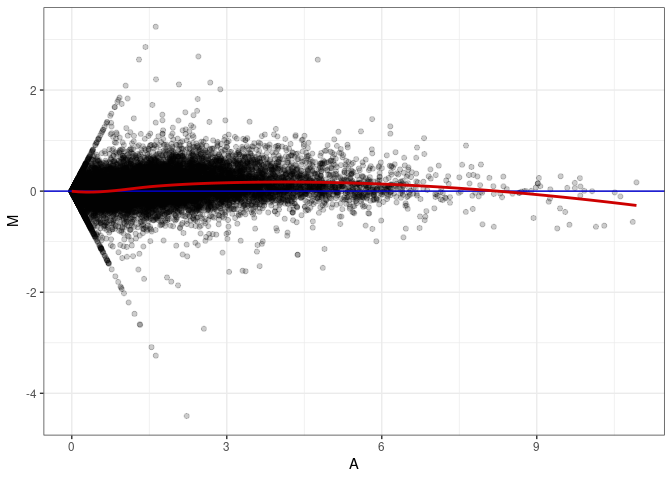
MA plot A039 - A003
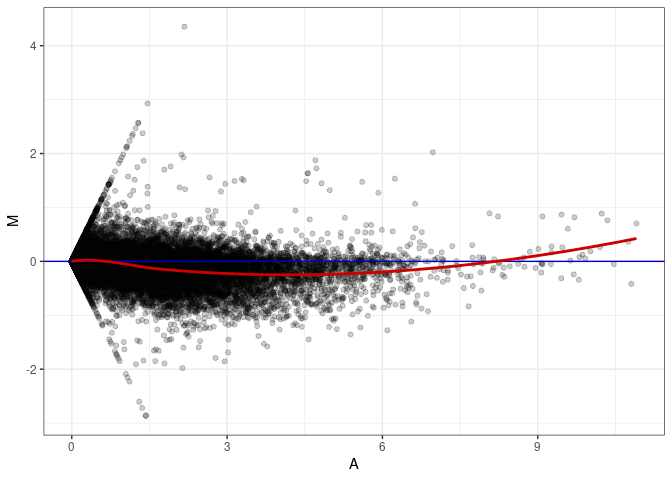
MA plot A039 - A039
MA plot A039 - A048
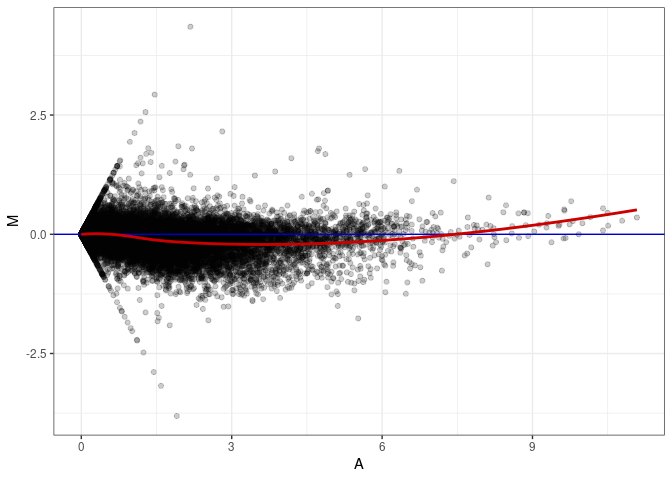
MA plot A039 - A084
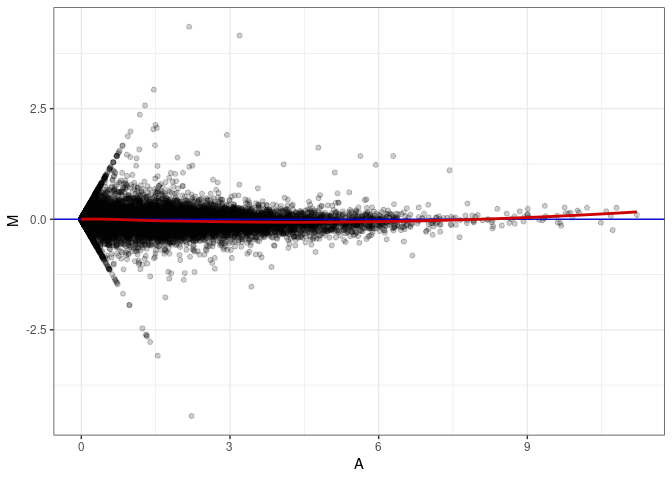
MA plot A048 - A003
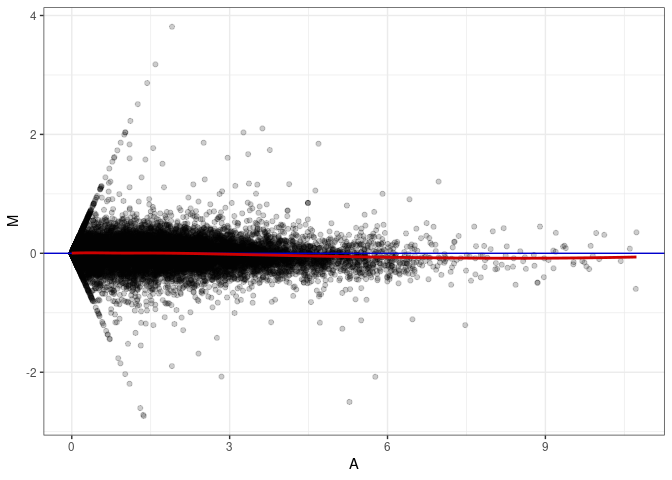
MA plot A048 - A039
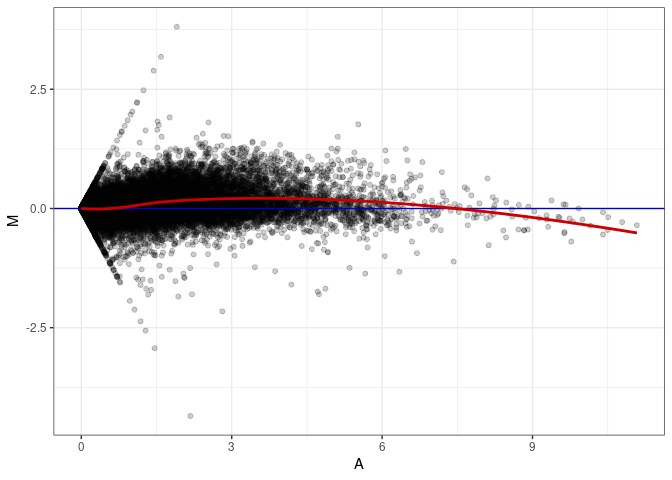
MA plot A048 - A048
MA plot A048 - A084
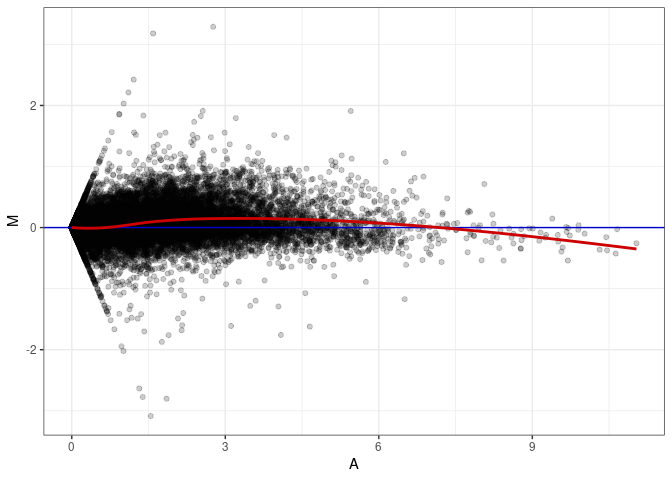
MA plot A084 - A003
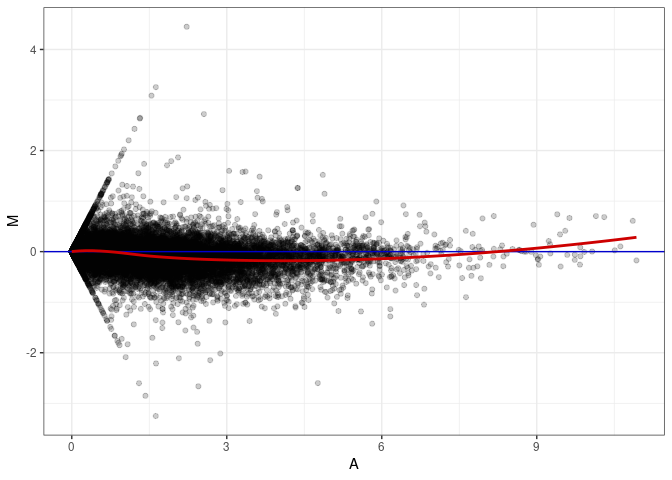
MA plot A084 - A039
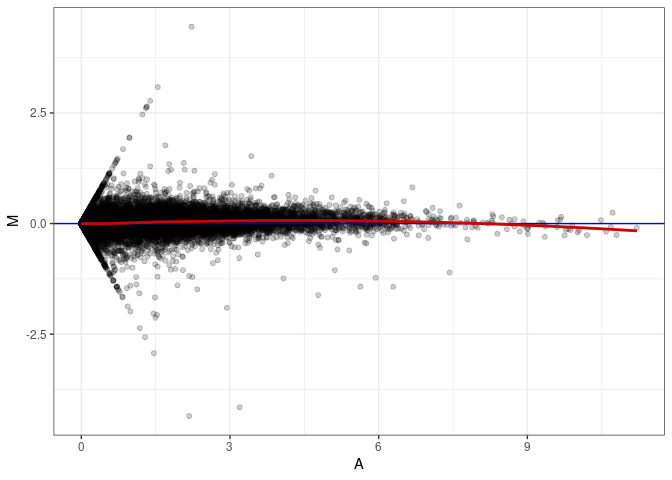
MA plot A084 - A048
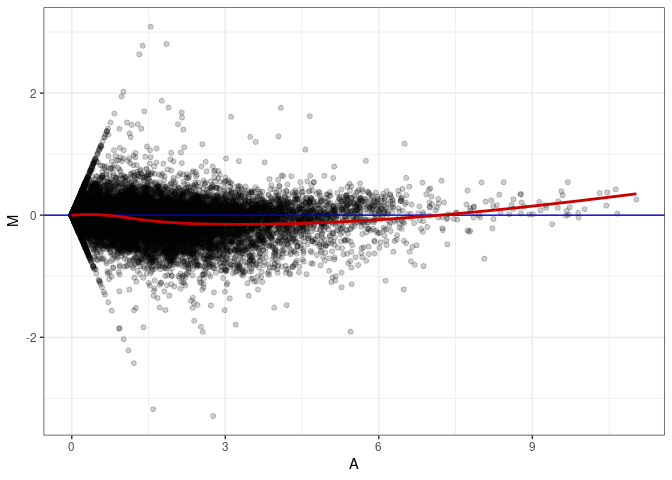
MA plot A084 - A084
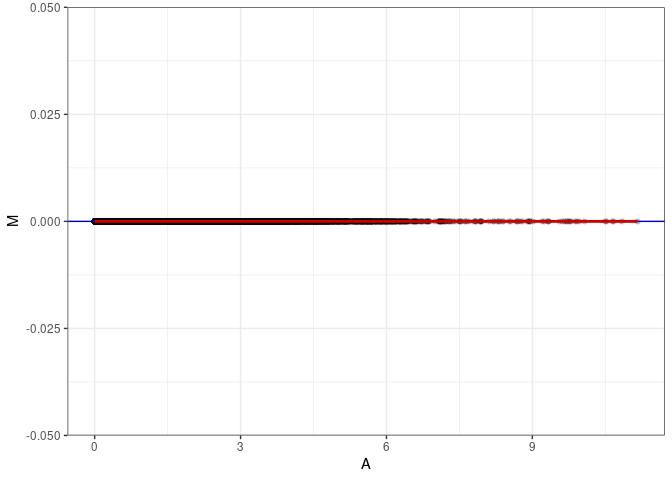
MA plot for Liver 2DG 96hrs
subdata <- log.tdata.FPKM.sample.info %>% rownames_to_column() %>% filter(Tissue =="Liver" & Treatment == "2DG" & Time == "96 hrs") %>% column_to_rownames()
subdata <- subdata[,-c(27237:27239)]
log.data.FPKM.sample.info <- t(subdata)
for(i in 1:ncol(log.data.FPKM.sample.info)){
for(j in 1:ncol(log.data.FPKM.sample.info)){
x = log.data.FPKM.sample.info[,i]
y = log.data.FPKM.sample.info[,j]
## M-values
M=x-y ## A-values
A = (x + y)/2
df = data.frame(A, M)
cat('###',"MA plot", colnames(log.data.FPKM.sample.info)[[i]], "-", colnames(log.data.FPKM.sample.info)[[j]] ,' \n')
p <- ggplot(df, aes(x = A, y = M)) + geom_point(size = 1.5, alpha = 1/5) +
geom_hline(color = "blue3", yintercept = 0) + stat_smooth(se = FALSE, method = "loess", color = "red3") + theme_bw()
print(p)
#print(htmltools::tagList(ggplotly(p)))
cat("\n \n")
}
}MA plot A012 - A012
MA plot A012 - A138
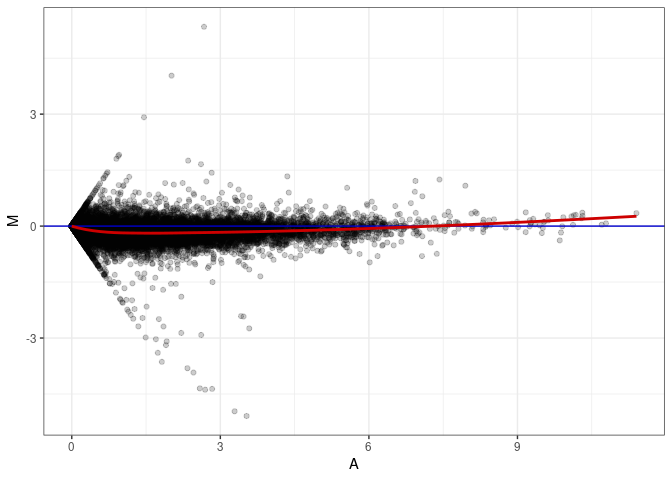
MA plot A012 - A147
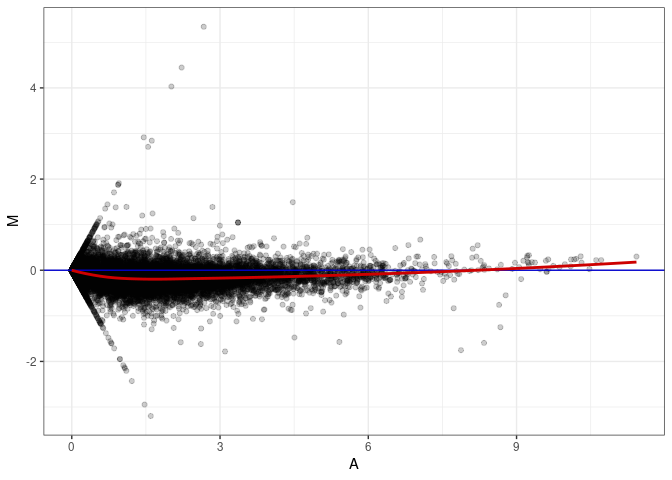
MA plot A012 - A165
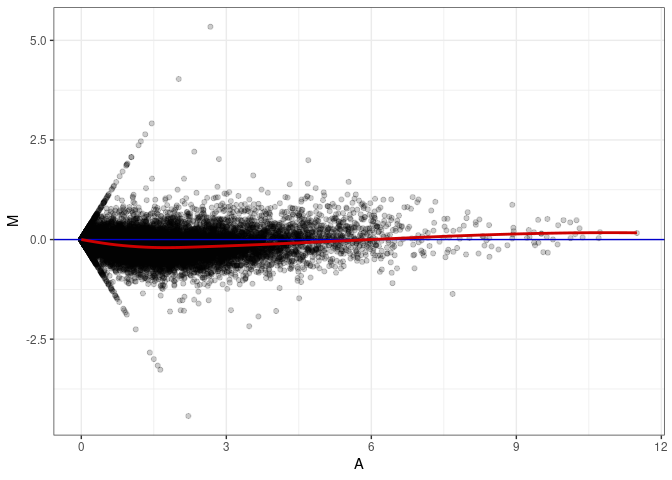
MA plot A138 - A012
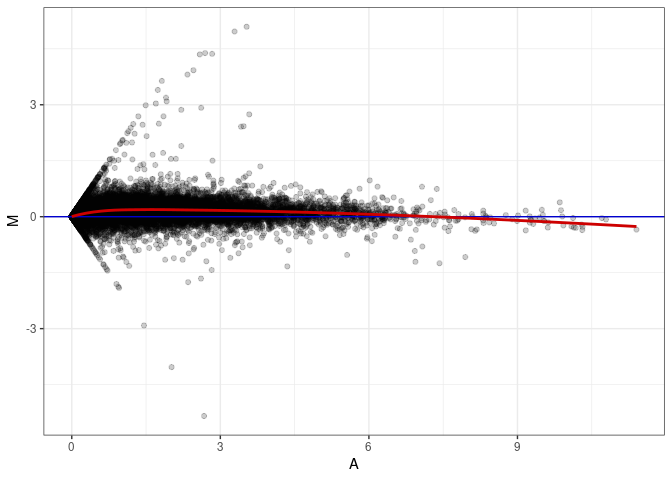
MA plot A138 - A138
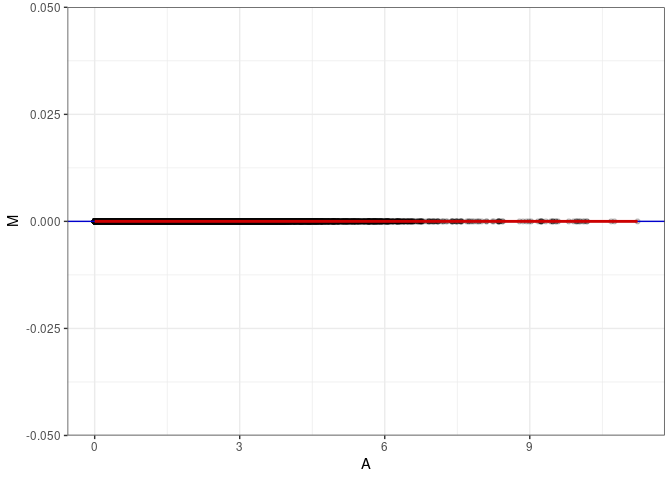
MA plot A138 - A147
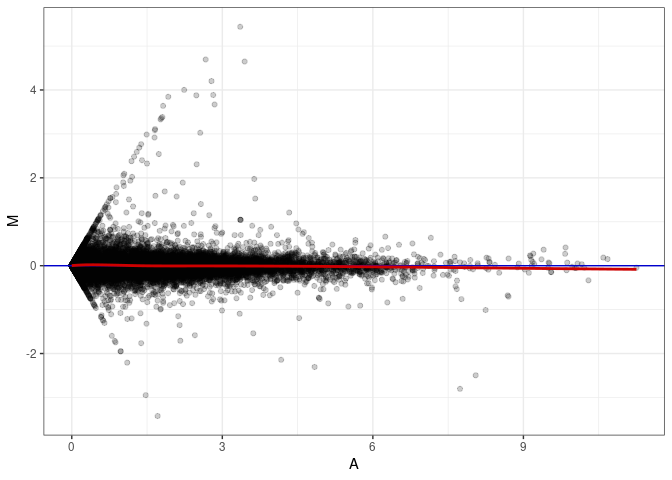
MA plot A138 - A165
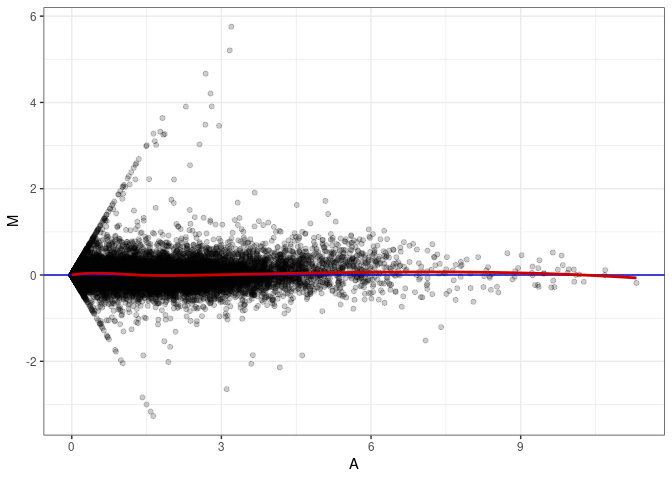
MA plot A147 - A012
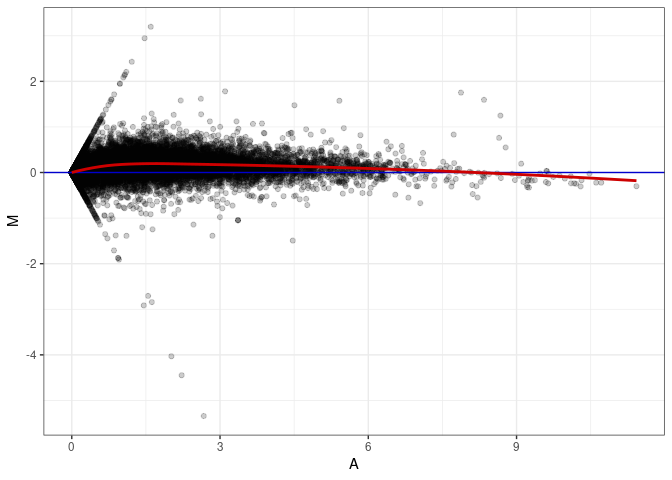
MA plot A147 - A138
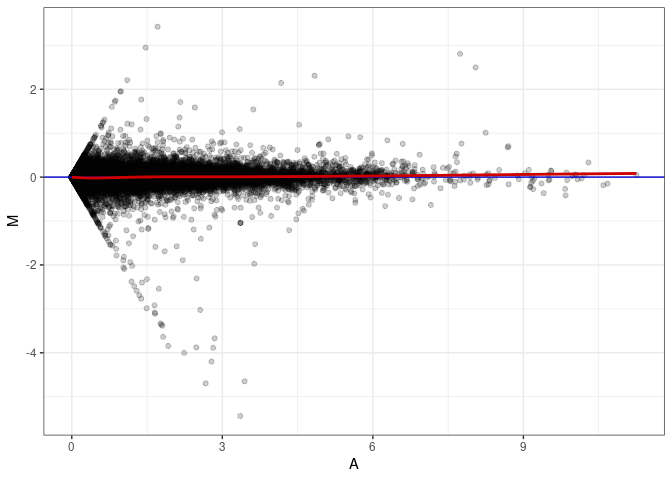
MA plot A147 - A147
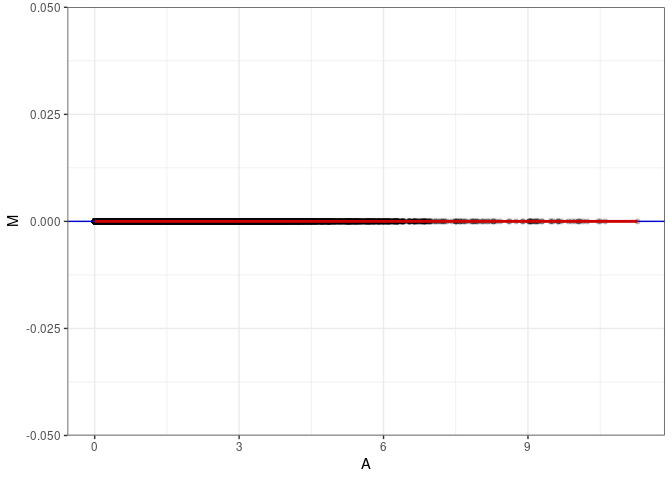
MA plot A147 - A165
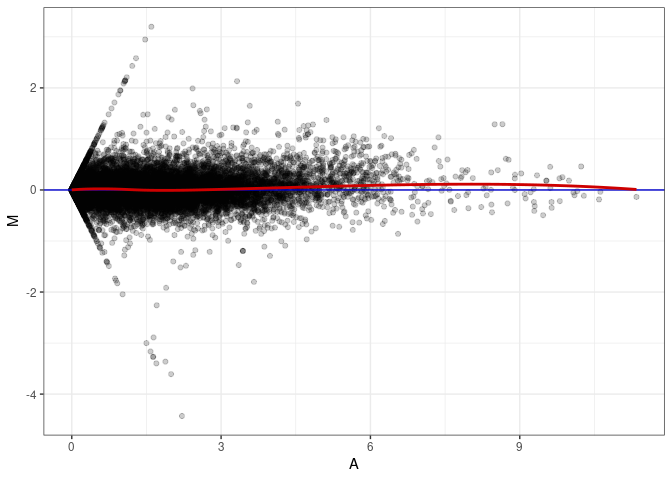
MA plot A165 - A012
MA plot A165 - A138
MA plot A165 - A147
MA plot A165 - A165
MA plot for Liver control 96hrs
subdata <- log.tdata.FPKM.sample.info %>% rownames_to_column() %>% filter(Tissue =="Liver" & Treatment == "None" & Time == "96 hrs") %>% column_to_rownames()
subdata <- subdata[,-c(27237:27239)]
log.data.FPKM.sample.info <- t(subdata)
for(i in 1:ncol(log.data.FPKM.sample.info)){
for(j in 1:ncol(log.data.FPKM.sample.info)){
x = log.data.FPKM.sample.info[,i]
y = log.data.FPKM.sample.info[,j]
## M-values
M=x-y ## A-values
A = (x + y)/2
df = data.frame(A, M)
cat('###',"MA plot", colnames(log.data.FPKM.sample.info)[[i]], "-", colnames(log.data.FPKM.sample.info)[[j]] ,' \n')
p <- ggplot(df, aes(x = A, y = M)) + geom_point(size = 1.5, alpha = 1/5) +
geom_hline(color = "blue3", yintercept = 0) + stat_smooth(se = FALSE, method = "loess", color = "red3") + theme_bw()
print(p)
#print(htmltools::tagList(ggplotly(p)))
cat("\n \n")
}
}MA plot A057 - A057
MA plot A057 - A066
MA plot A057 - A075
MA plot A057 - A111
MA plot A066 - A057
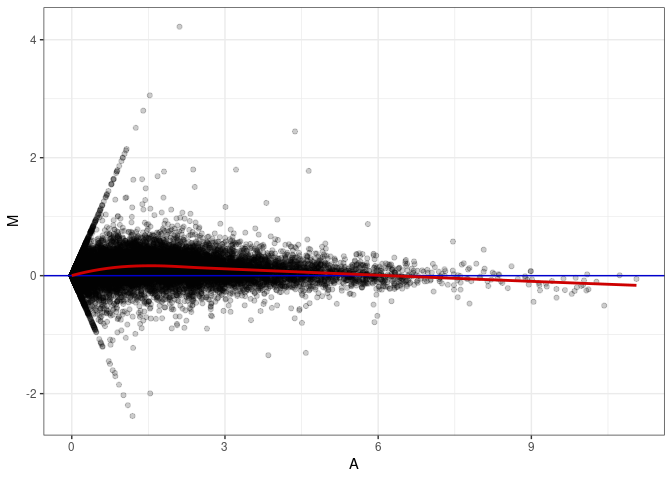
MA plot A066 - A066
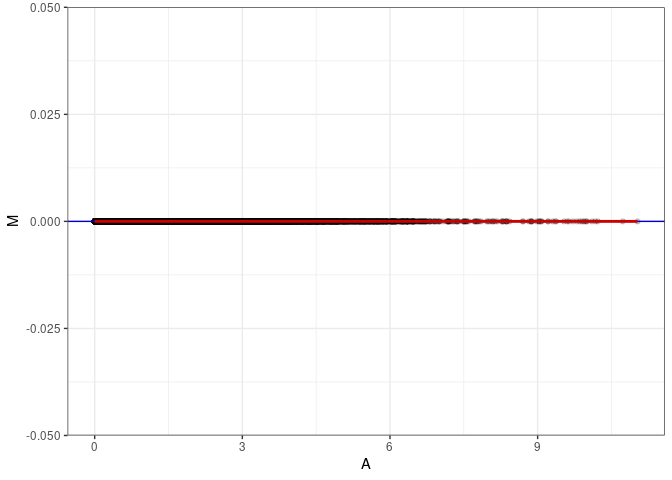
MA plot A066 - A075
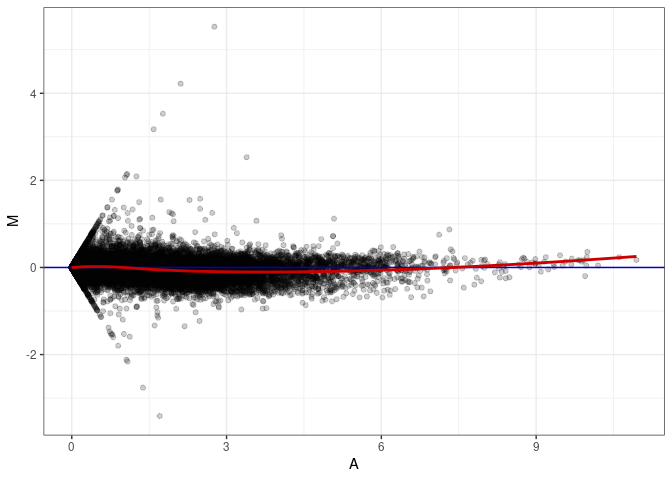
MA plot A066 - A111
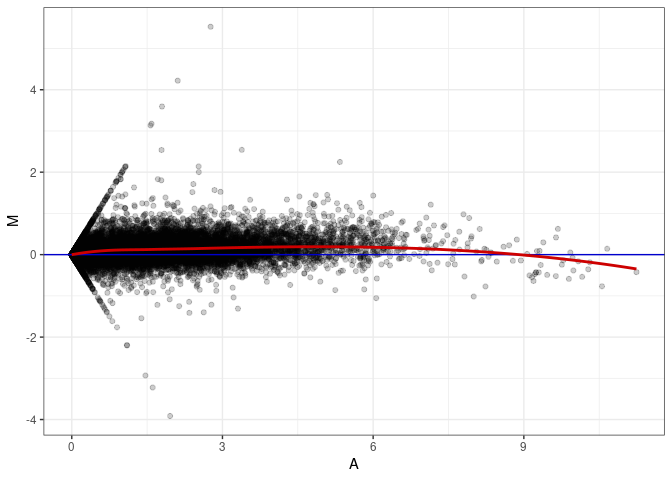
MA plot A075 - A057
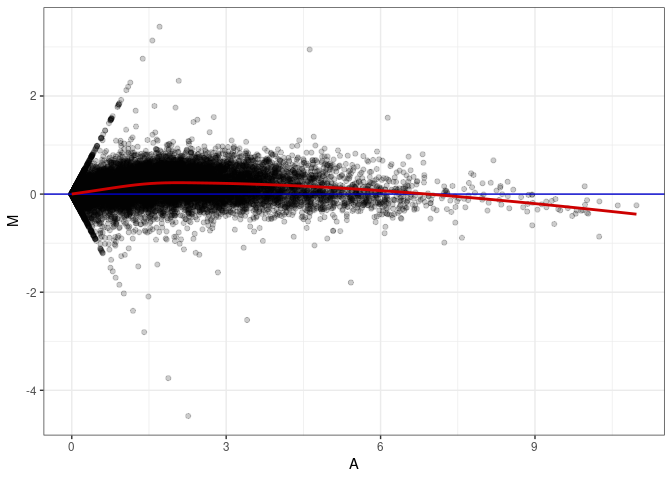
MA plot A075 - A066
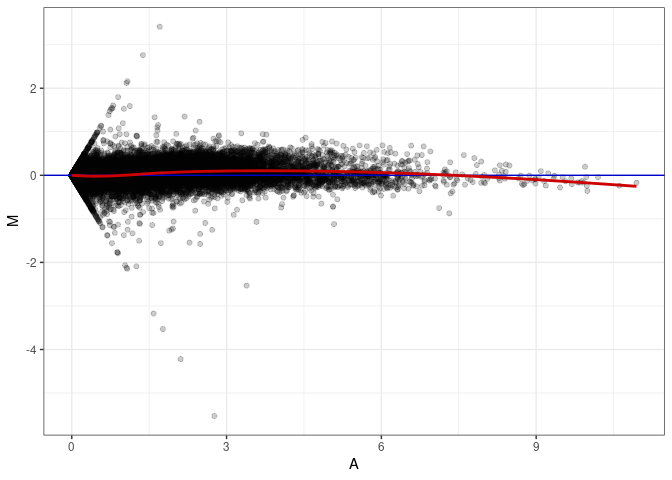
MA plot A075 - A075
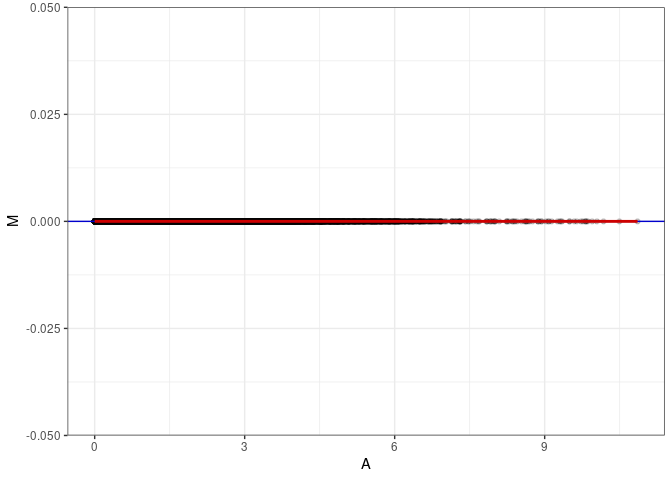
MA plot A075 - A111
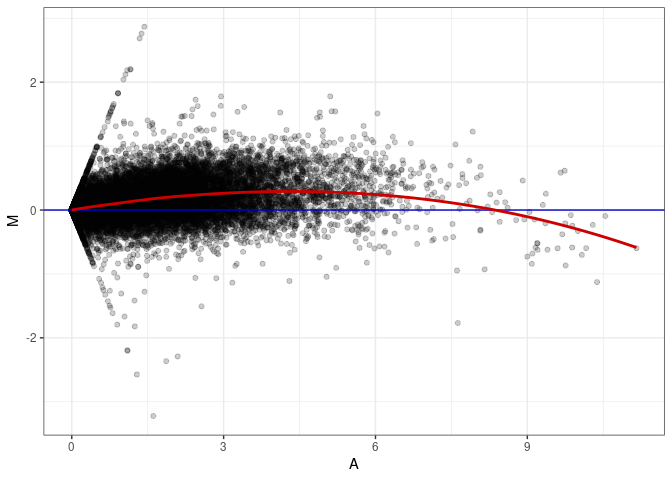
MA plot A111 - A057
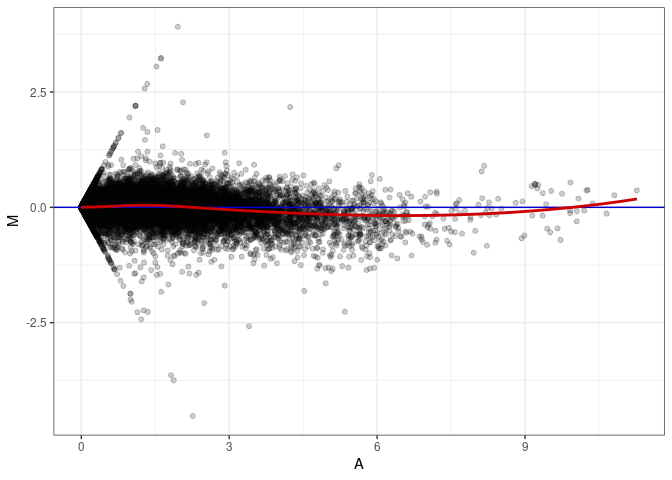
MA plot A111 - A066
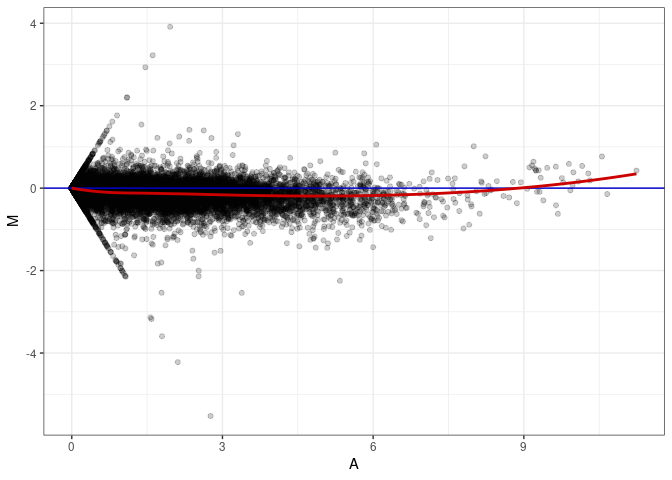
MA plot A111 - A075
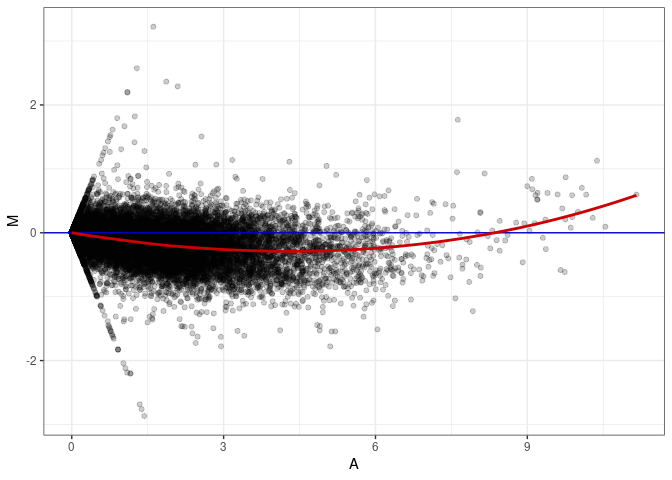
MA plot A111 - A111
MA plot for Heart 2DG 4wks
subdata <- log.tdata.FPKM.sample.info %>% rownames_to_column() %>% filter(Tissue =="Heart" & Treatment == "2DG" & Time == "4 wks") %>% column_to_rownames()
subdata <- subdata[,-c(27237:27239)]
log.data.FPKM.sample.info <- t(subdata)
for(i in 1:ncol(log.data.FPKM.sample.info)){
for(j in 1:ncol(log.data.FPKM.sample.info)){
x = log.data.FPKM.sample.info[,i]
y = log.data.FPKM.sample.info[,j]
## M-values
M=x-y ## A-values
A = (x + y)/2
df = data.frame(A, M)
if(i != j){
cat('###',"MA plot", colnames(log.data.FPKM.sample.info)[[i]], "-", colnames(log.data.FPKM.sample.info)[[j]] ,' \n')
p <- ggplot(df, aes(x = A, y = M)) + geom_point(size = 1.5, alpha = 1/5) +
geom_hline(color = "blue3", yintercept = 0) + stat_smooth(se = FALSE, method = "loess", color = "red3") + theme_bw()
print(p)
#print(htmltools::tagList(ggplotly(p)))
cat("\n \n")
}
}
}MA plot A022 - A031
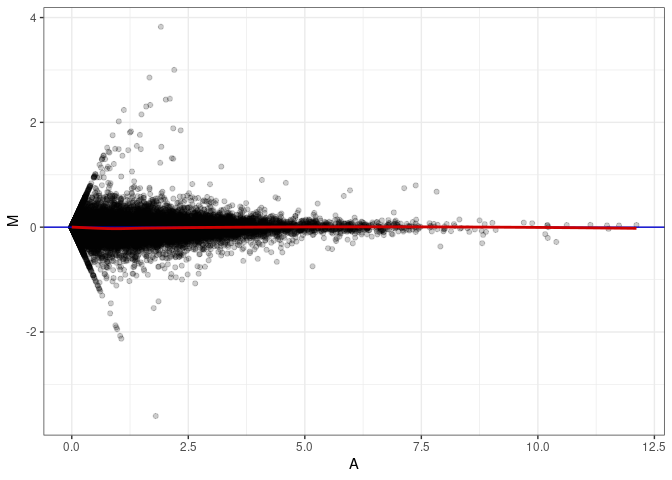
MA plot A022 - A094
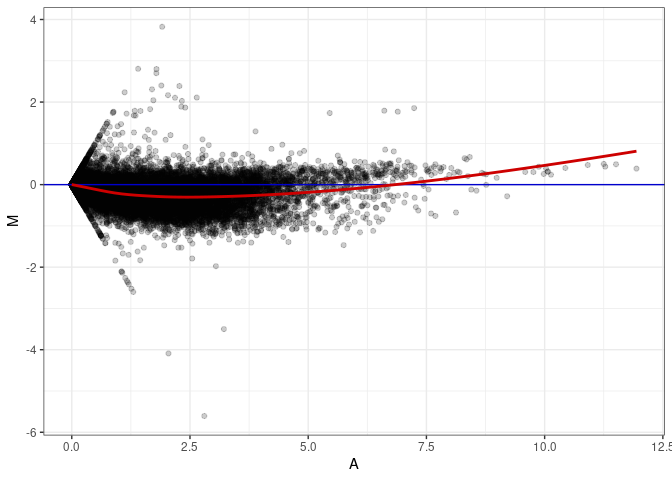
MA plot A022 - A175
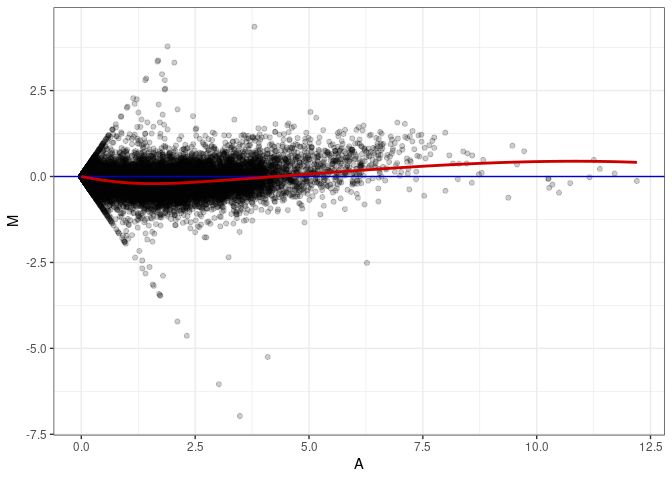
MA plot A031 - A022
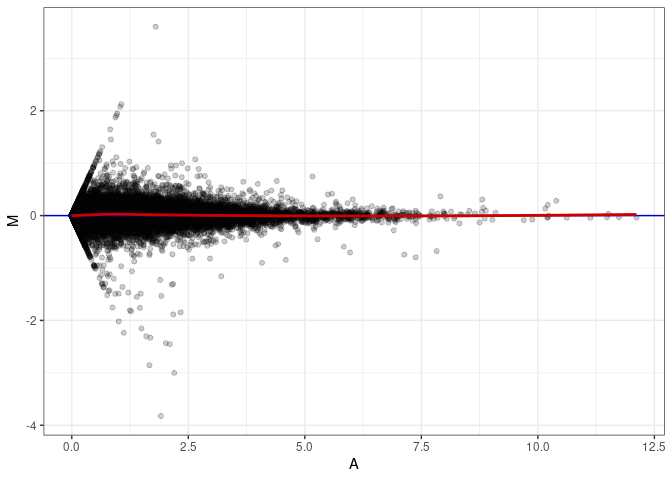
MA plot A031 - A094
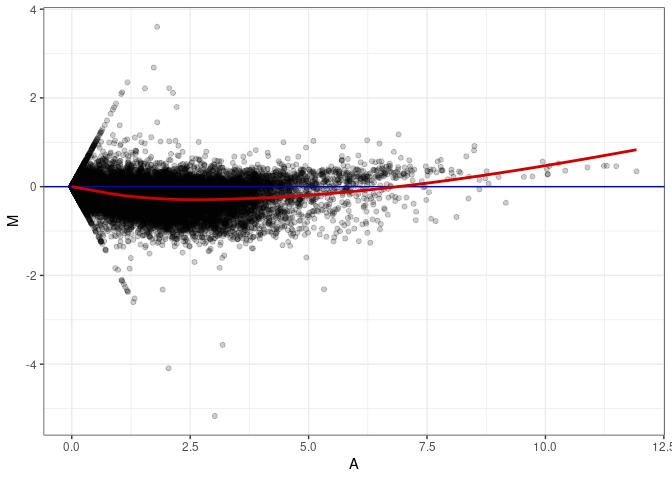
MA plot A031 - A175
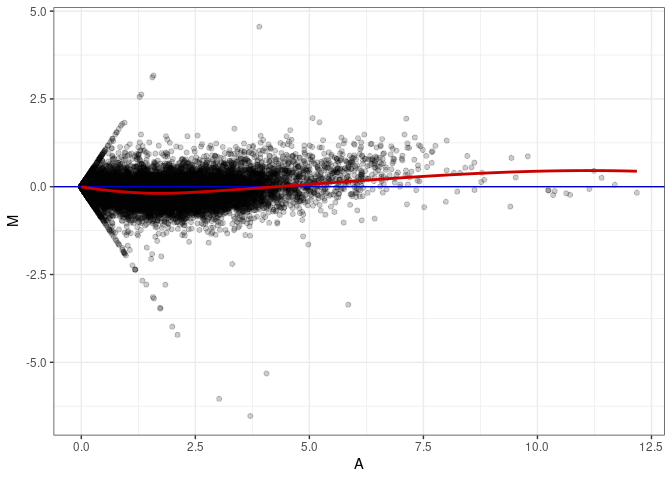
MA plot A094 - A022
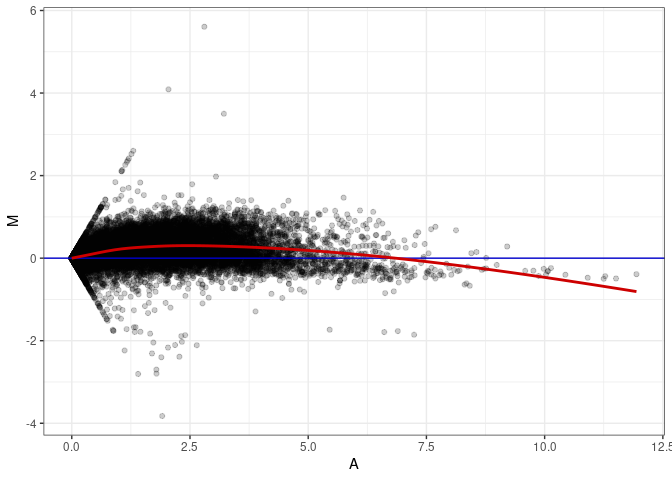
MA plot A094 - A031
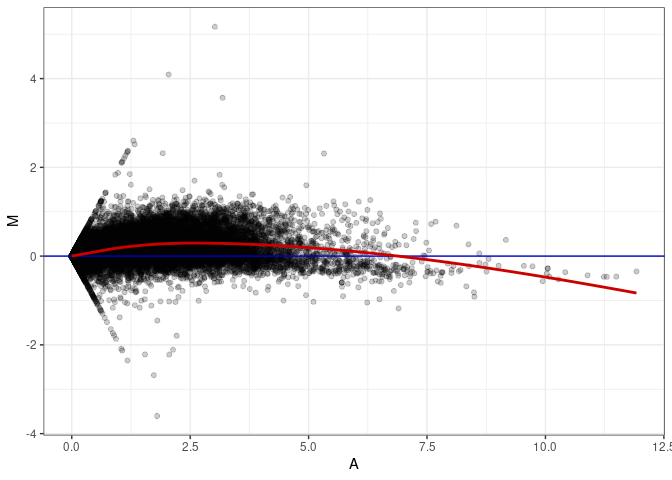
MA plot A094 - A175
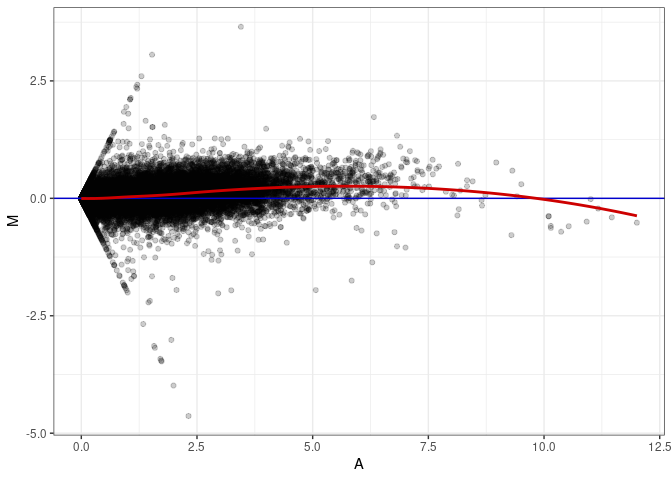
MA plot A175 - A022
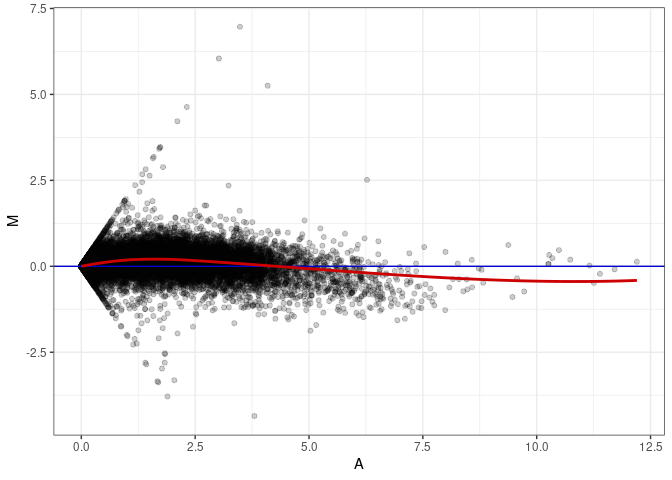
MA plot A175 - A031
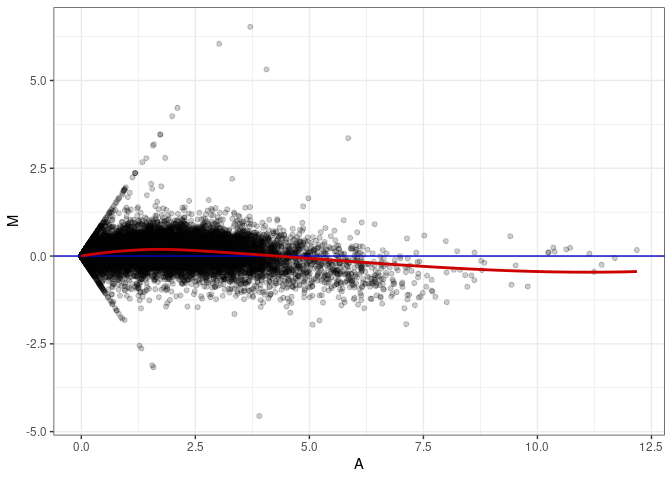
MA plot A175 - A094
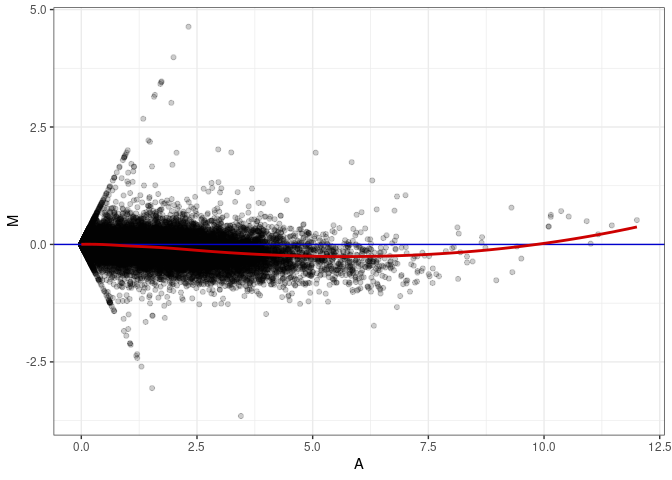
MA plot for Heart control 4wks
subdata <- log.tdata.FPKM.sample.info %>% rownames_to_column() %>% filter(Tissue =="Heart" & Treatment == "None" & Time == "4 wks") %>% column_to_rownames()
subdata <- subdata[,-c(27237:27239)]
log.data.FPKM.sample.info <- t(subdata)
for(i in 1:ncol(log.data.FPKM.sample.info)){
for(j in 1:ncol(log.data.FPKM.sample.info)){
x = log.data.FPKM.sample.info[,i]
y = log.data.FPKM.sample.info[,j]
## M-values
M=x-y ## A-values
A = (x + y)/2
df = data.frame(A, M)
cat('###',"MA plot", colnames(log.data.FPKM.sample.info)[[i]], "-", colnames(log.data.FPKM.sample.info)[[j]] ,' \n')
p <- ggplot(df, aes(x = A, y = M)) + geom_point(size = 1.5, alpha = 1/5) +
geom_hline(color = "blue3", yintercept = 0) + stat_smooth(se = FALSE, method = "loess", color = "red3") + theme_bw()
print(p)
#print(htmltools::tagList(ggplotly(p)))
cat("\n \n")
}
}MA plot A004 - A004
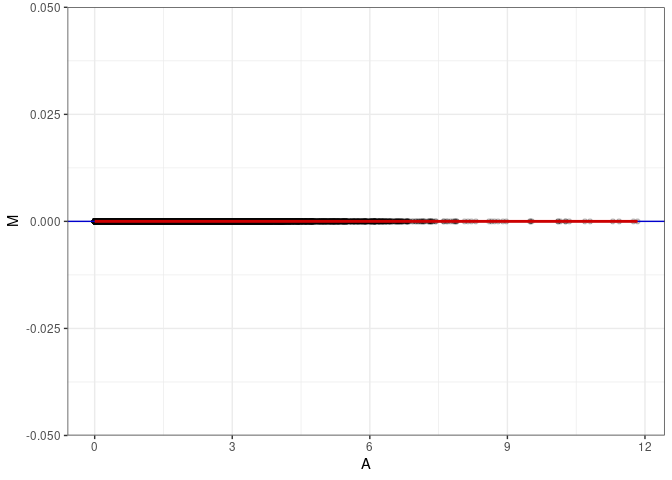
MA plot A004 - A040
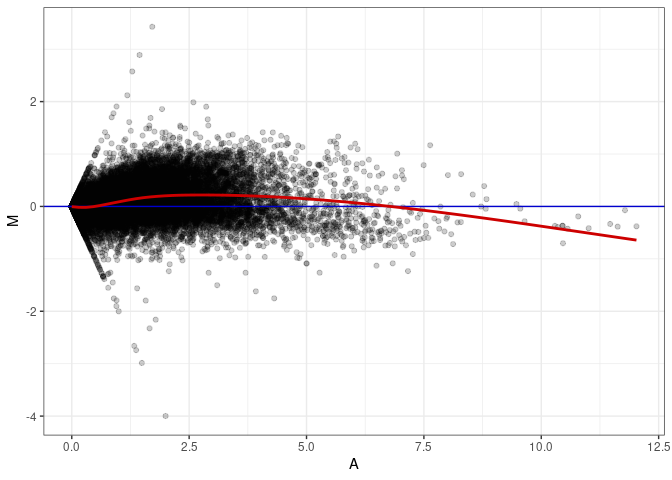
MA plot A004 - A049
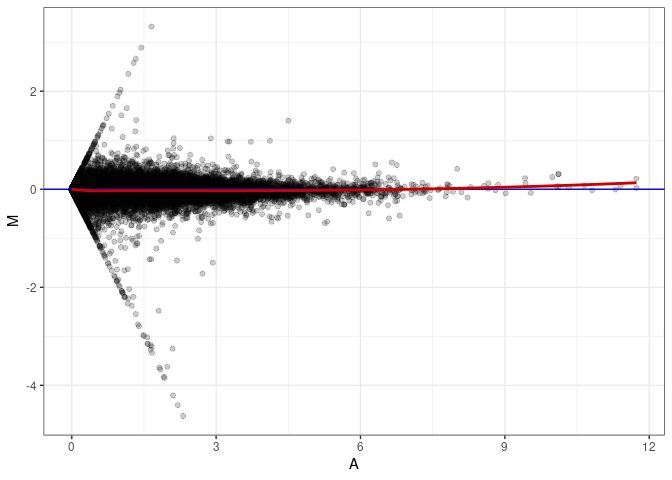
MA plot A004 - A085
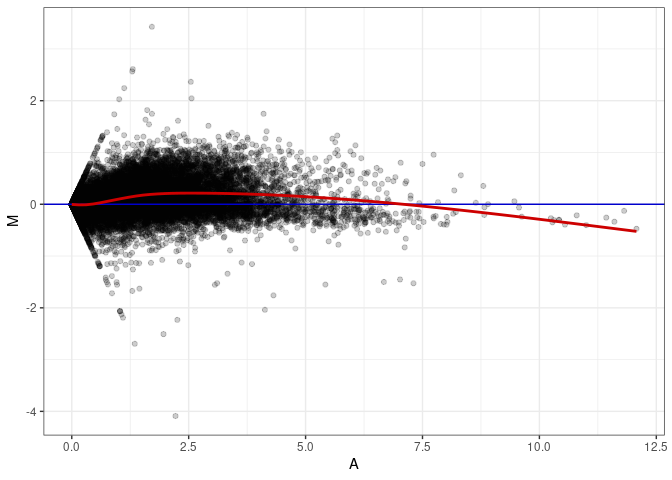
MA plot A040 - A004
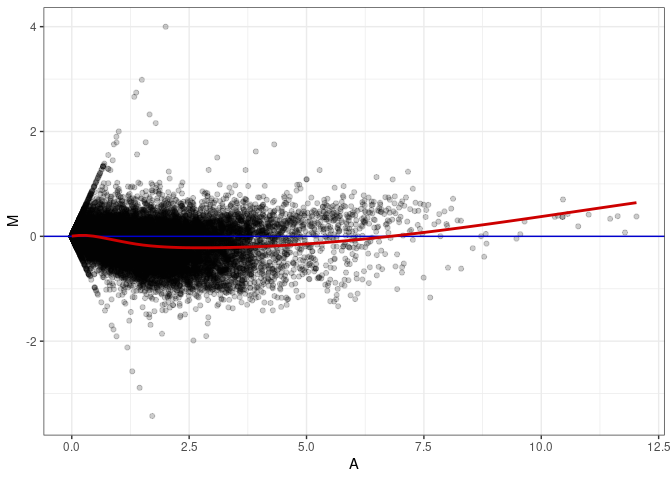
MA plot A040 - A040
MA plot A040 - A049
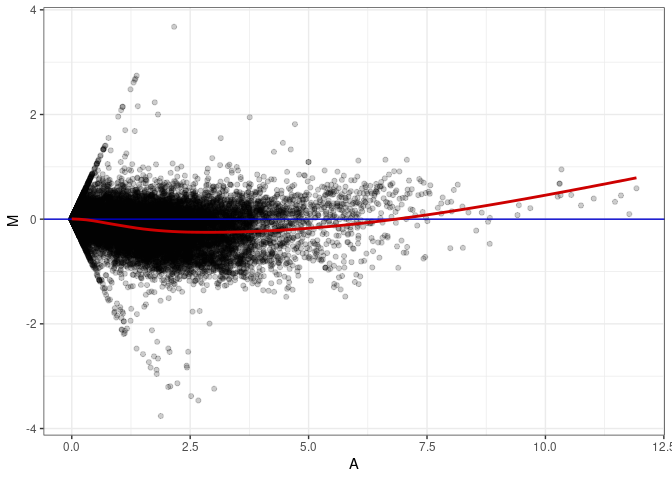
MA plot A040 - A085
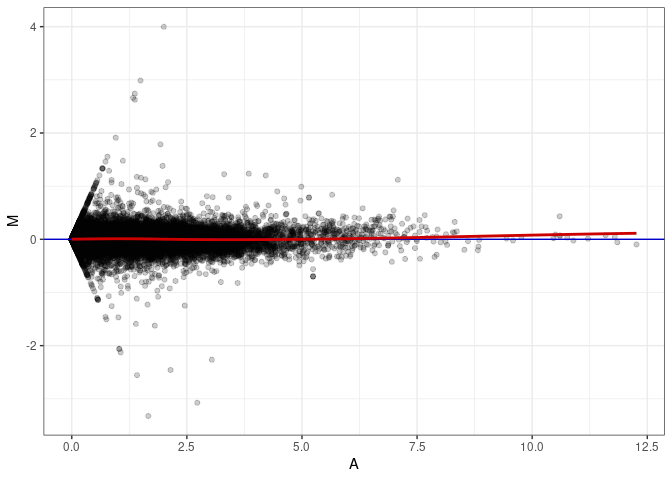
MA plot A049 - A004
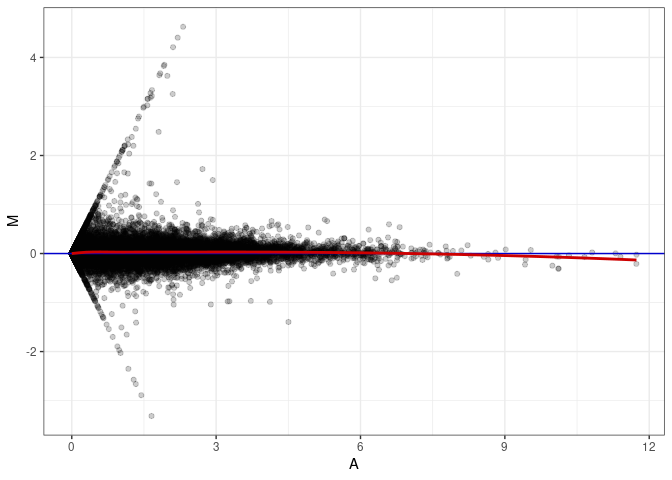
MA plot A049 - A040
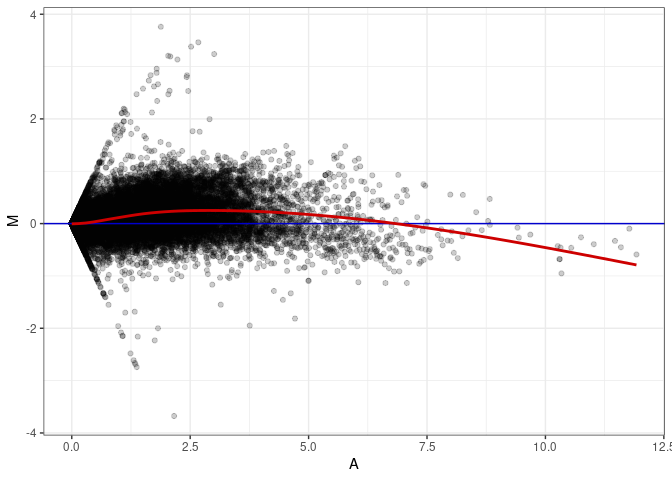
MA plot A049 - A049
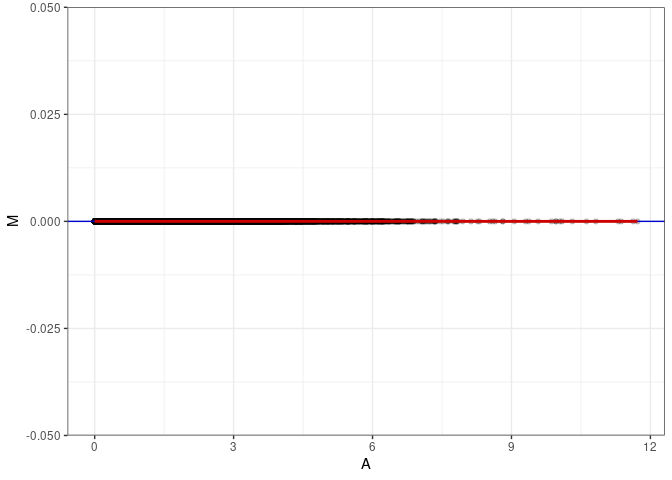
MA plot A049 - A085
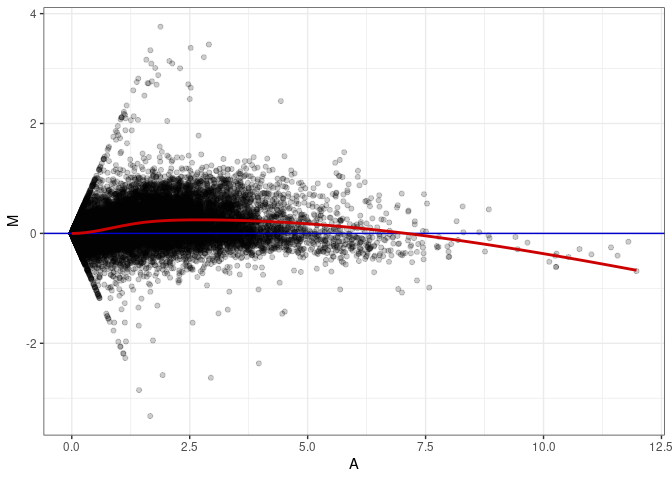
MA plot A085 - A004
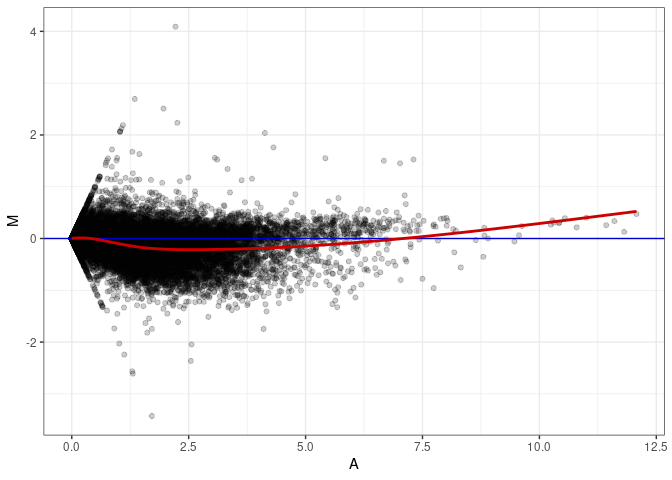
MA plot A085 - A040
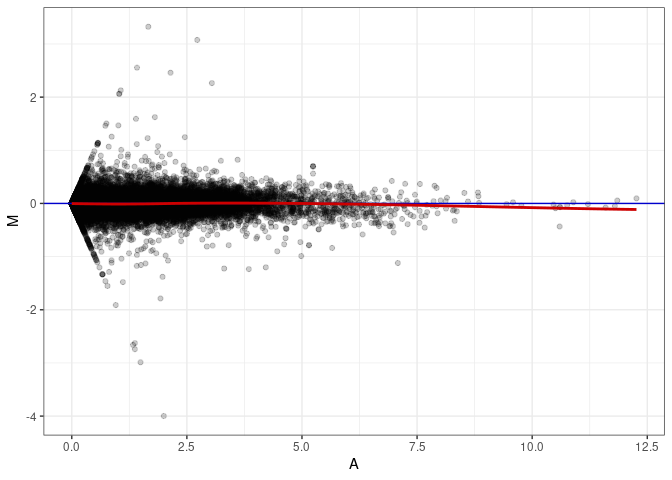
MA plot A085 - A049
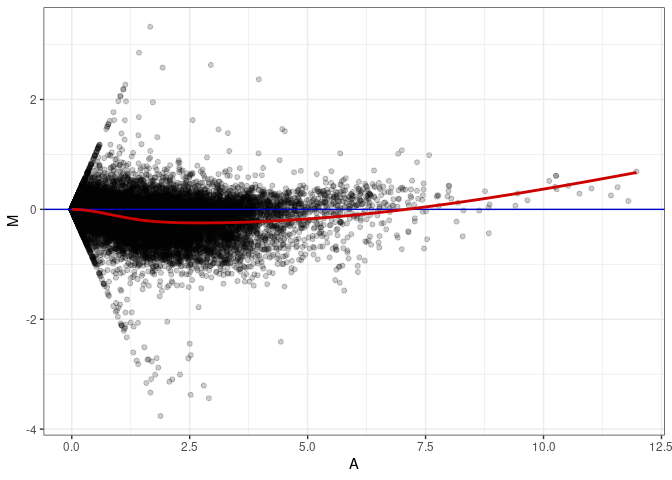
MA plot A085 - A085
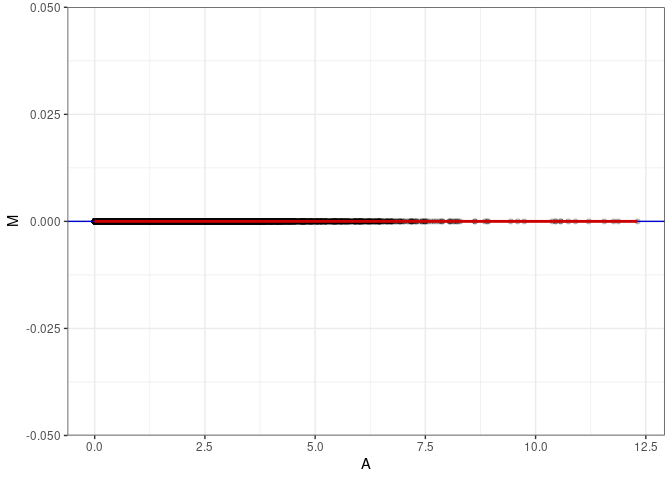
MA plot for Heart 2DG 96hrs
subdata <- log.tdata.FPKM.sample.info %>% rownames_to_column() %>% filter(Tissue =="Heart" & Treatment == "2DG" & Time == "96 hrs") %>% column_to_rownames()
subdata <- subdata[,-c(27237:27239)]
log.data.FPKM.sample.info <- t(subdata)
for(i in 1:ncol(log.data.FPKM.sample.info)){
for(j in 1:ncol(log.data.FPKM.sample.info)){
x = log.data.FPKM.sample.info[,i]
y = log.data.FPKM.sample.info[,j]
## M-values
M=x-y ## A-values
A = (x + y)/2
df = data.frame(A, M)
cat('###',"MA plot", colnames(log.data.FPKM.sample.info)[[i]], "-", colnames(log.data.FPKM.sample.info)[[j]] ,' \n')
p <- ggplot(df, aes(x = A, y = M)) + geom_point(size = 1.5, alpha = 1/5) +
geom_hline(color = "blue3", yintercept = 0) + stat_smooth(se = FALSE, method = "loess", color = "red3") + theme_bw()
print(p)
#print(htmltools::tagList(ggplotly(p)))
cat("\n \n")
}
}MA plot A013 - A013
MA plot A013 - A139
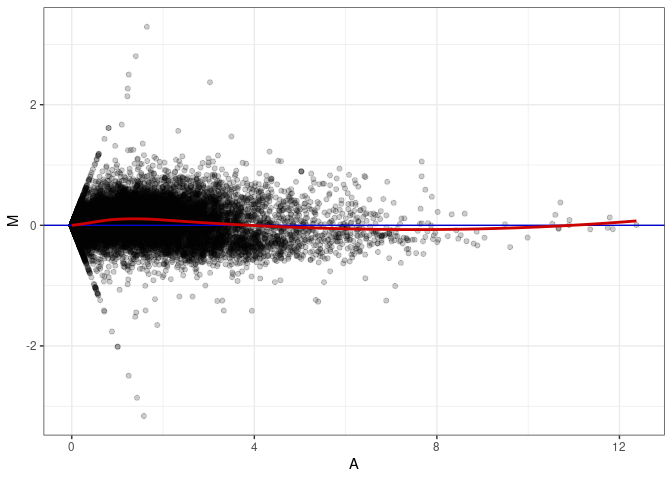
MA plot A013 - A148
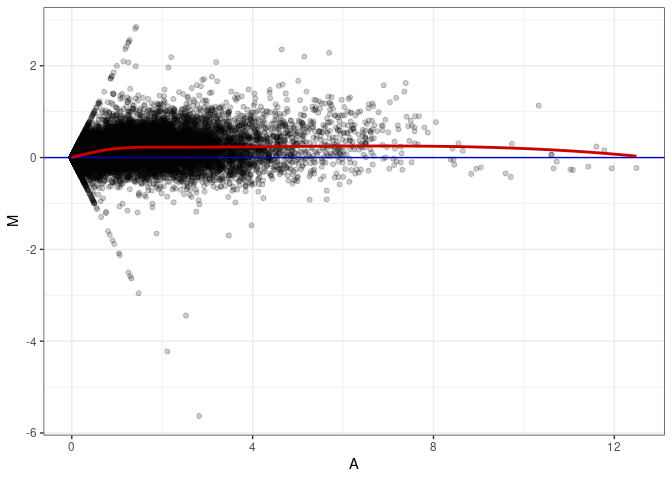
MA plot A013 - A166
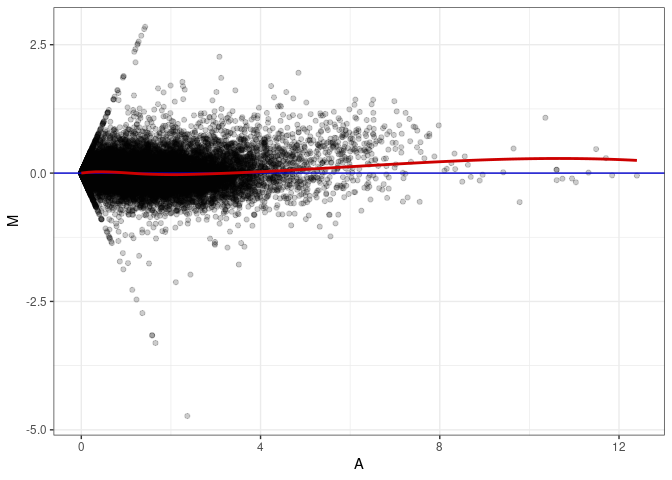
MA plot A139 - A013
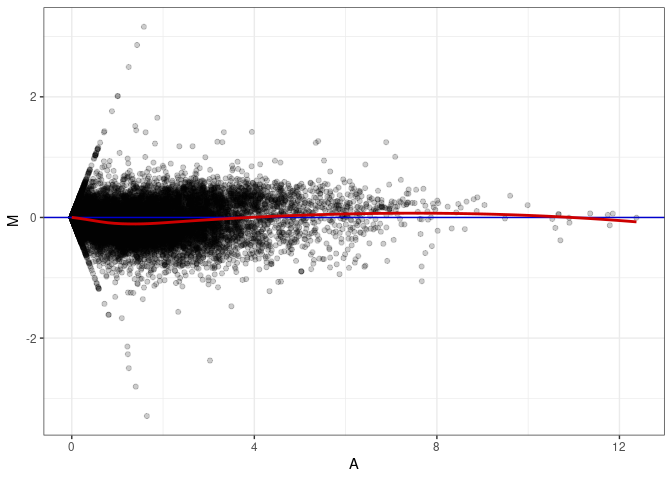
MA plot A139 - A139
MA plot A139 - A148
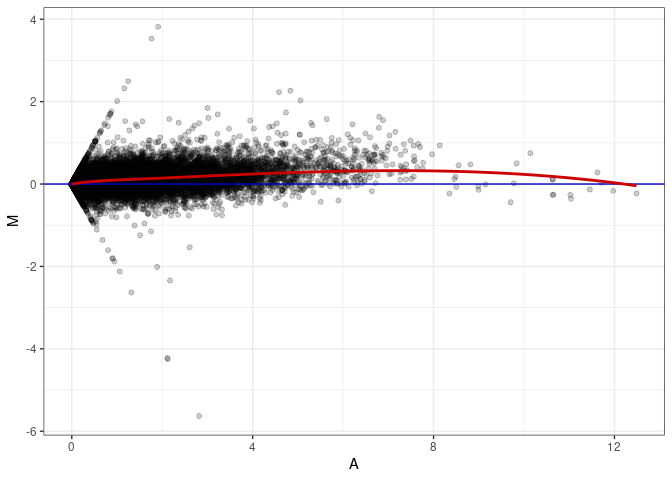
MA plot A139 - A166
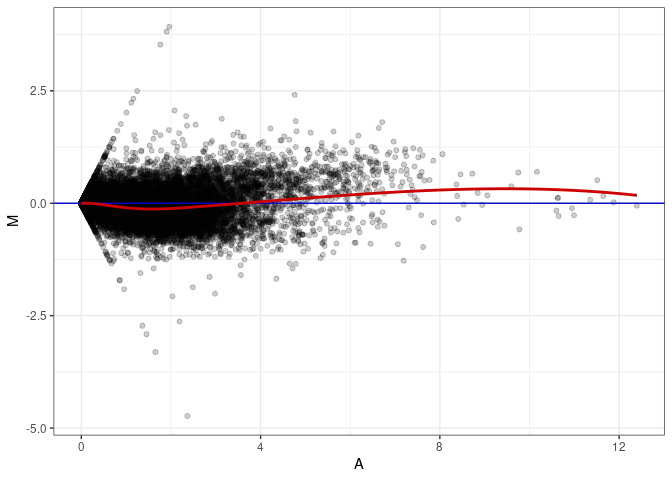
MA plot A148 - A013
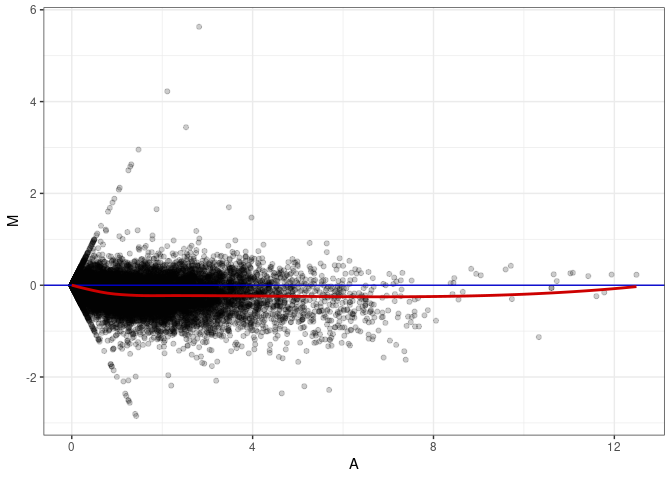
MA plot A148 - A139
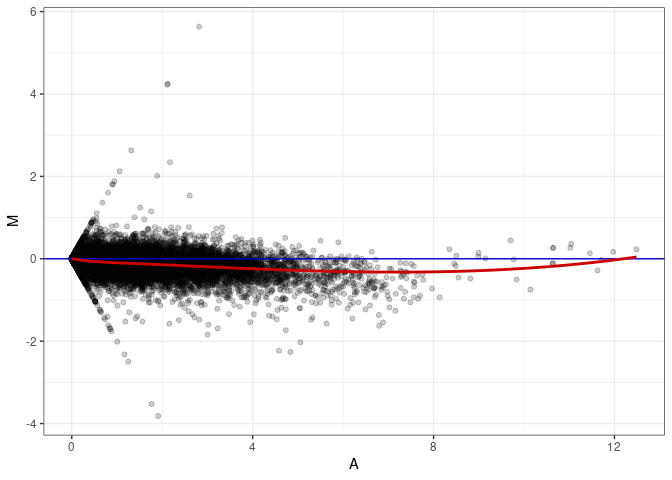
MA plot A148 - A148
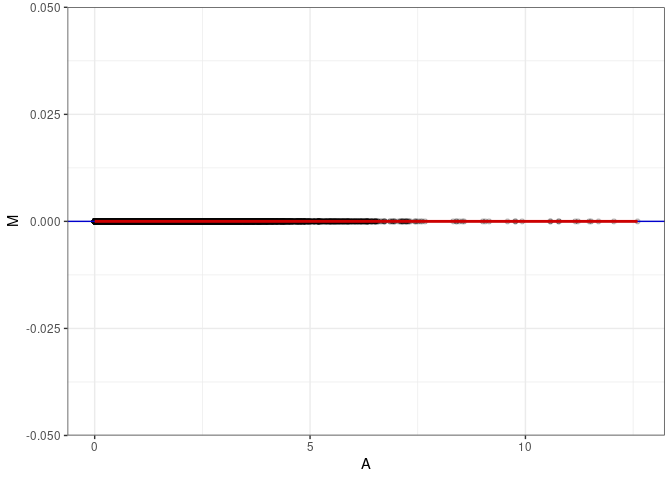
MA plot A148 - A166
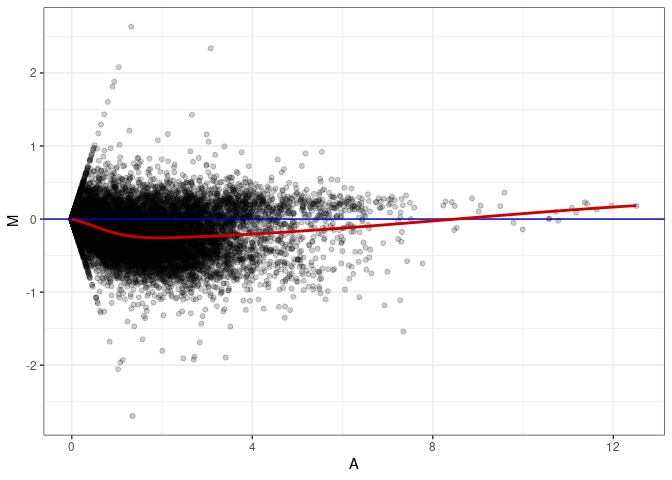
MA plot A166 - A013
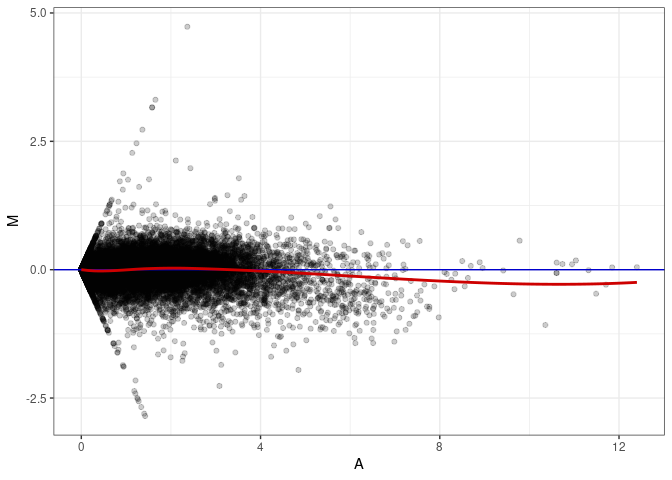
MA plot A166 - A139
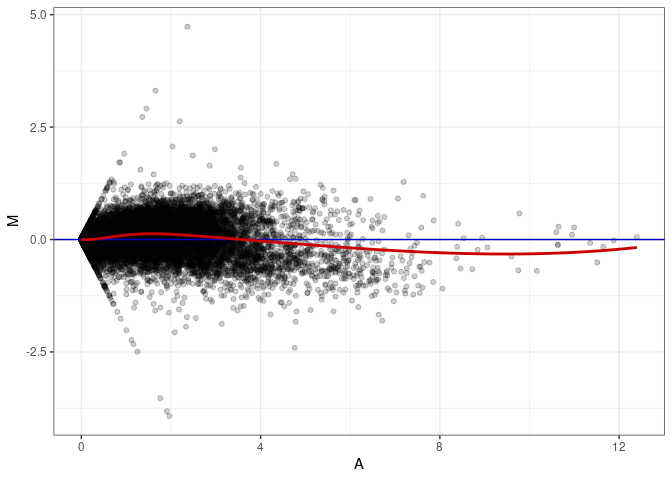
MA plot A166 - A148
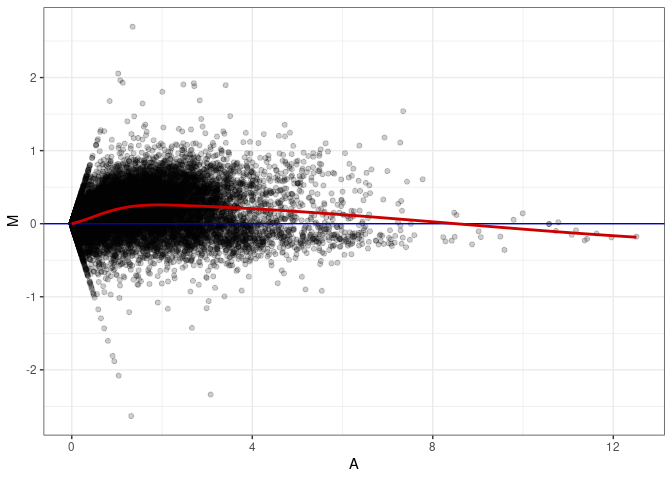
MA plot A166 - A166
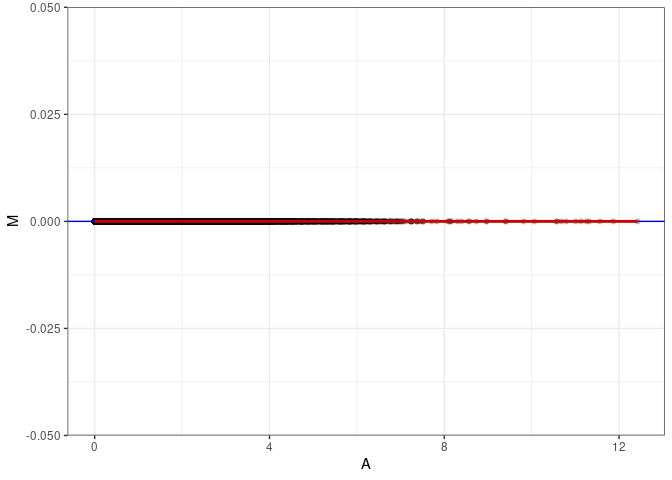
MA plot for Heart control 96hrs
subdata <- log.tdata.FPKM.sample.info %>% rownames_to_column() %>% filter(Tissue =="Heart" & Treatment == "None" & Time == "96 hrs") %>% column_to_rownames()
subdata <- subdata[,-c(27237:27239)]
log.data.FPKM.sample.info <- t(subdata)
for(i in 1:ncol(log.data.FPKM.sample.info)){
for(j in 1:ncol(log.data.FPKM.sample.info)){
x = log.data.FPKM.sample.info[,i]
y = log.data.FPKM.sample.info[,j]
##M-values
M=x-y ##A-values
A = (x + y)/2
df = data.frame(A, M)
cat('###',"MA plot", colnames(log.data.FPKM.sample.info)[[i]], "-", colnames(log.data.FPKM.sample.info)[[j]] ,' \n')
p <- ggplot(df, aes(x = A, y = M)) + geom_point(size = 1.5, alpha = 1/5) +
geom_hline(color = "blue3", yintercept = 0) + stat_smooth(se = FALSE, method = "loess", color = "red3") + theme_bw()
print(p)
#print(htmltools::tagList(ggplotly(p)))
cat("\n \n")
}
}MA plot A058 - A058
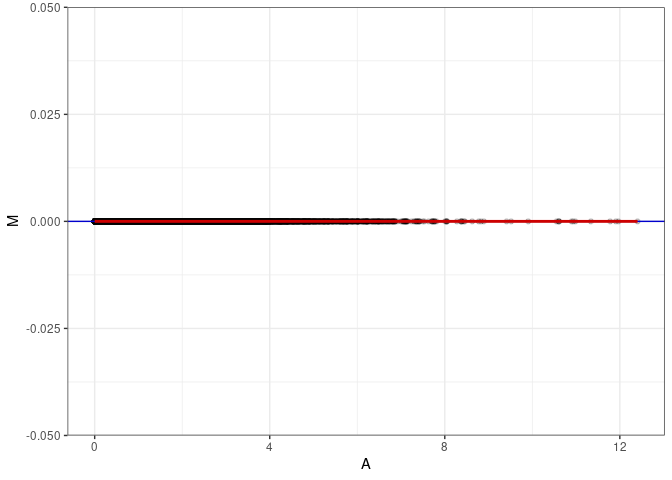
MA plot A058 - A067
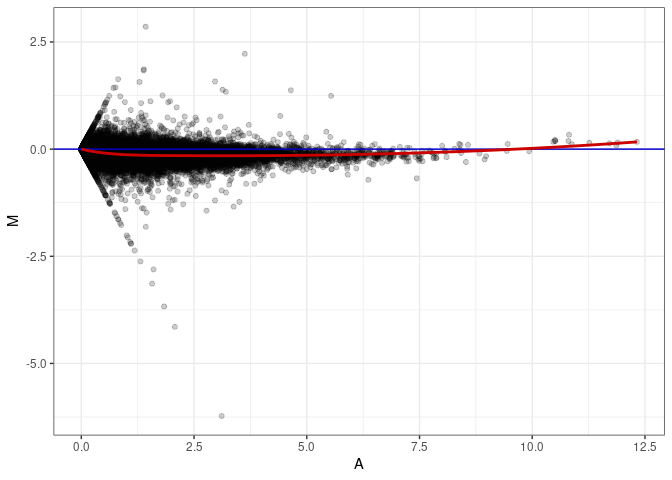
MA plot A058 - A076
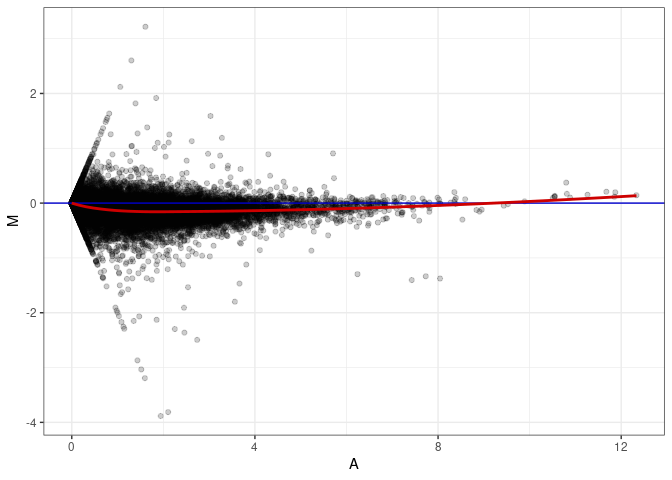
MA plot A058 - A112
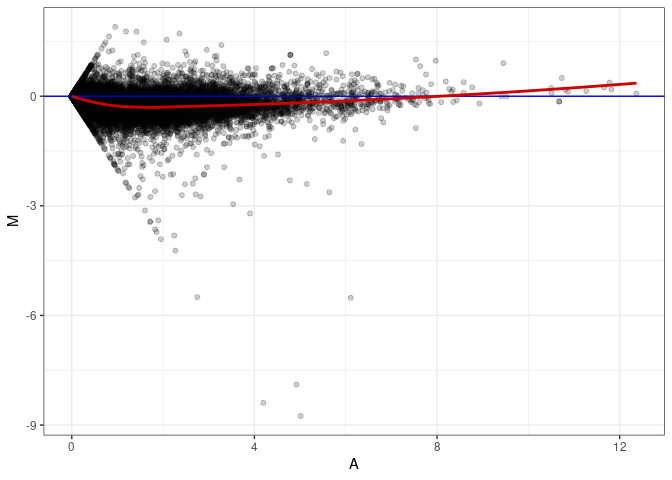
MA plot A067 - A058
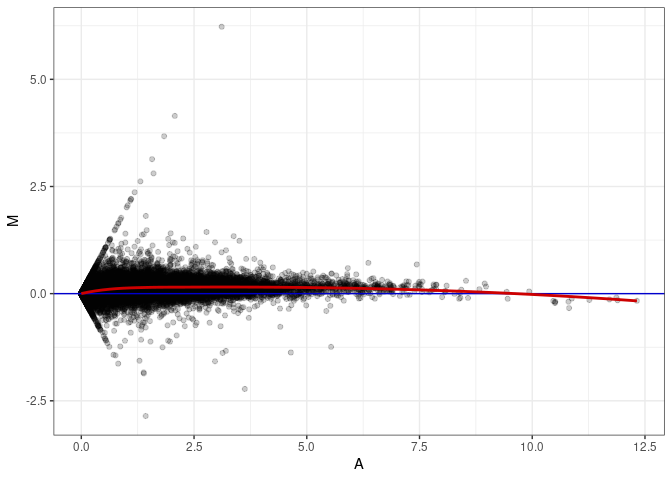
MA plot A067 - A067
MA plot A067 - A076
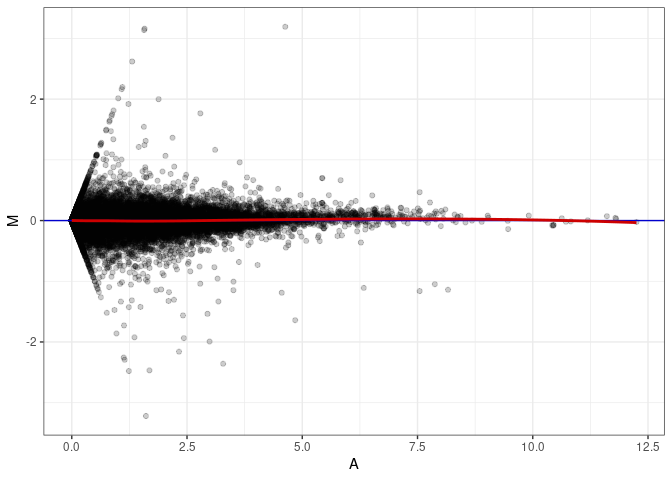
MA plot A067 - A112
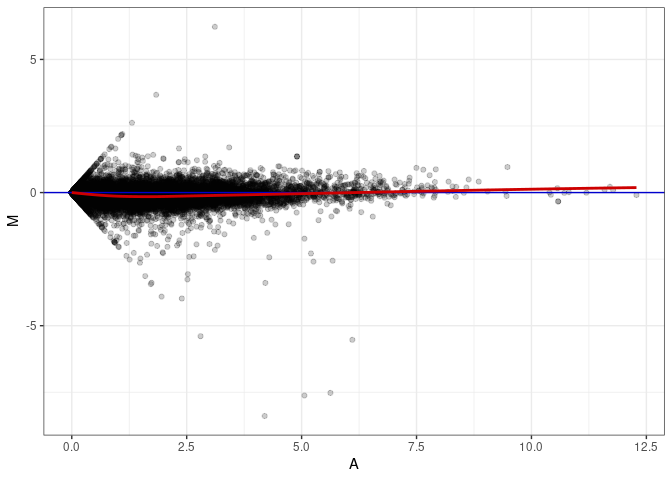
MA plot A076 - A058
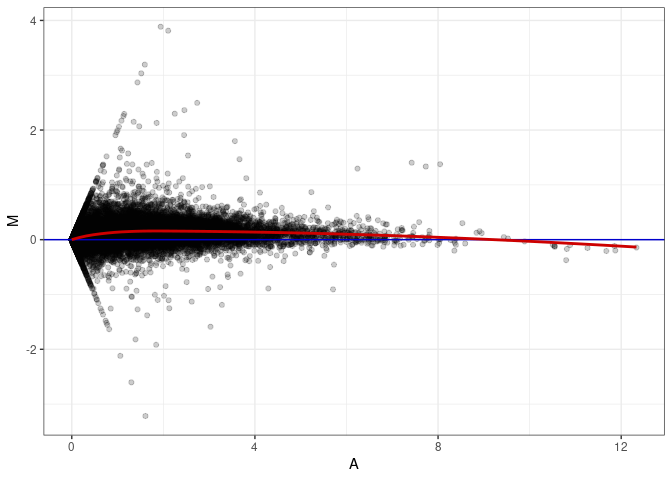
MA plot A076 - A067
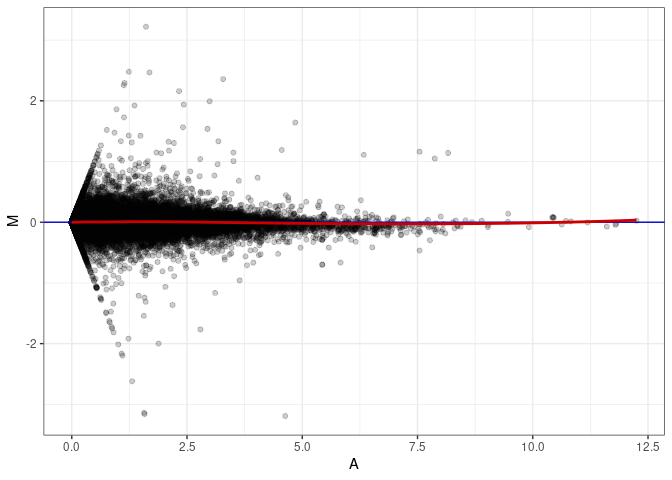
MA plot A076 - A076
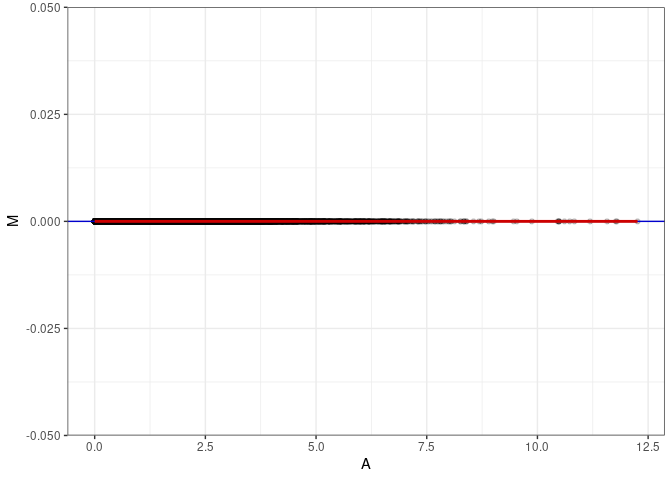
MA plot A076 - A112
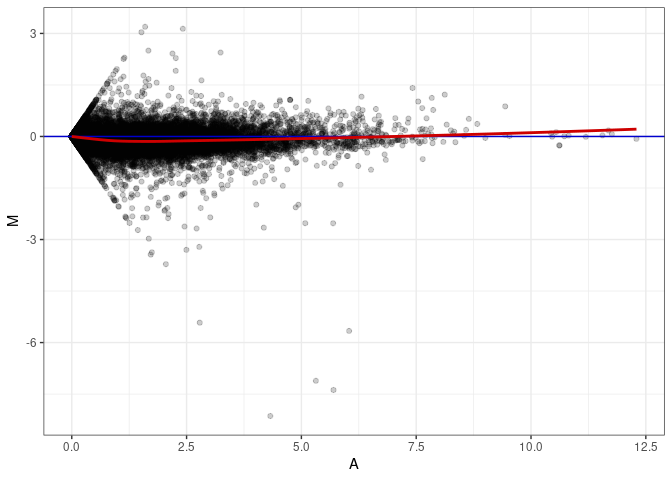
MA plot A112 - A058
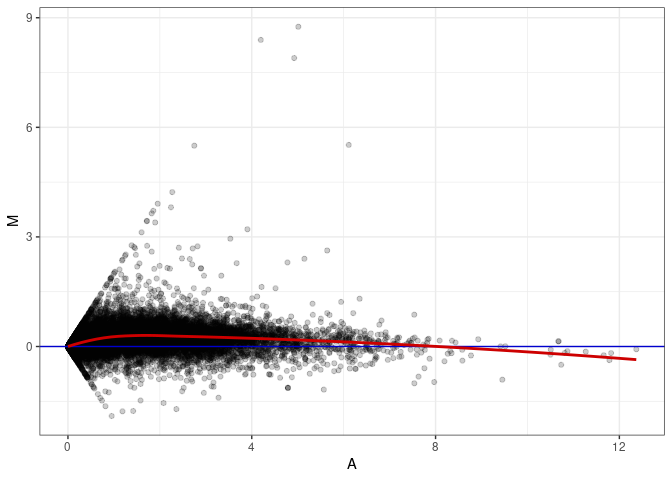
MA plot A112 - A067
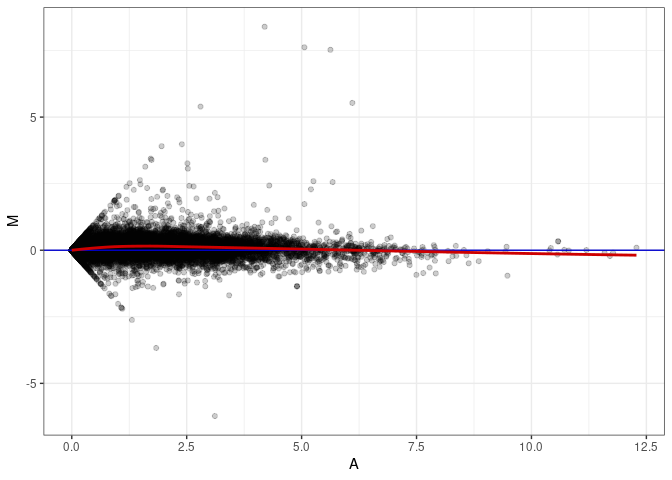
MA plot A112 - A076
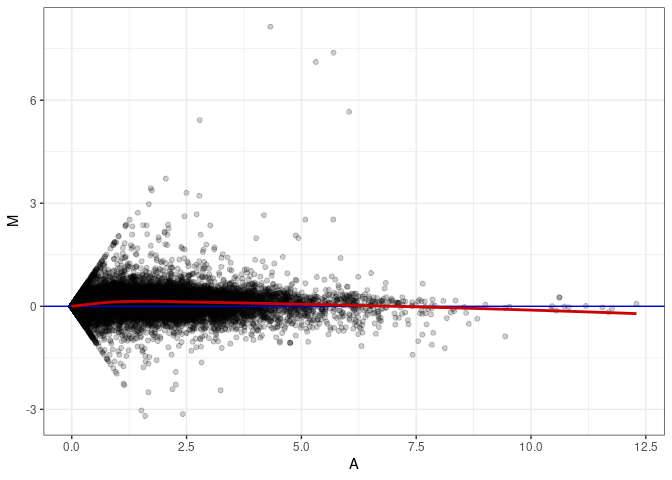
MA plot A112 - A112
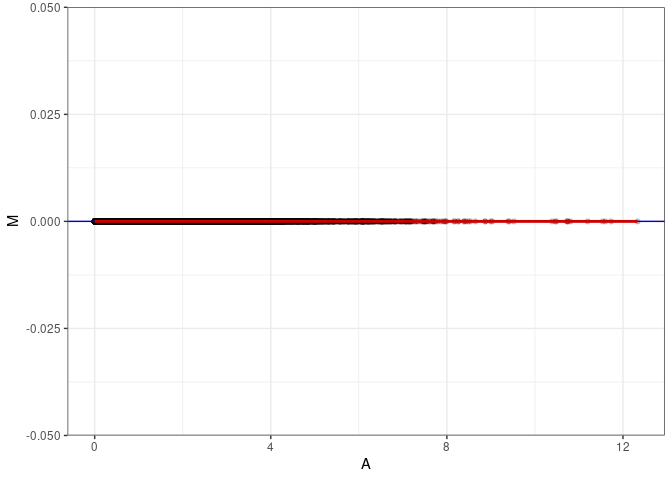
MA plot for Small Intestine 2DG 4wks
subdata <- log.tdata.FPKM.sample.info %>% rownames_to_column() %>% filter(Tissue =="Small Intestine" & Treatment == "2DG" & Time == "4 wks") %>% column_to_rownames()
subdata <- subdata[,-c(27237:27239)]
log.data.FPKM.sample.info <- t(subdata)
for(i in 1:ncol(log.data.FPKM.sample.info)){
for(j in 1:ncol(log.data.FPKM.sample.info)){
x = log.data.FPKM.sample.info[,i]
y = log.data.FPKM.sample.info[,j]
## M-values
M=x-y ## A-values
A = (x + y)/2
df = data.frame(A, M)
cat('###',"MA plot", colnames(log.data.FPKM.sample.info)[[i]], "-", colnames(log.data.FPKM.sample.info)[[j]] ,' \n')
p <- ggplot(df, aes(x = A, y = M)) + geom_point(size = 1.5, alpha = 1/5) +
geom_hline(color = "blue3", yintercept = 0) + stat_smooth(se = FALSE, method = "loess", color = "red3") + theme_bw()
print(p)
#print(htmltools::tagList(ggplotly(p)))
cat("\n \n")
}
}MA plot A024 - A024
MA plot A024 - A033
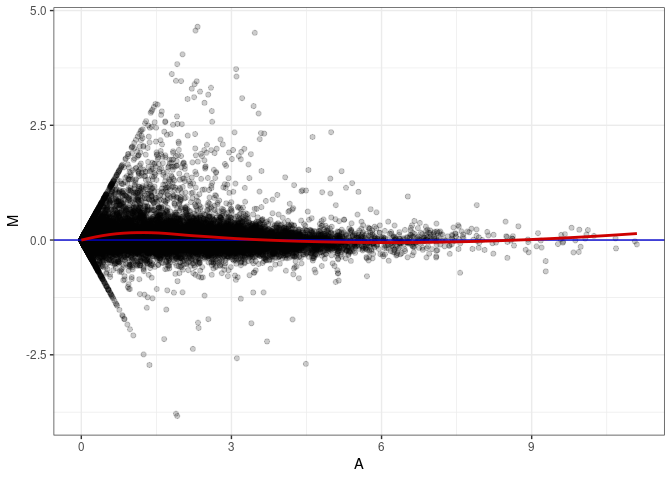
MA plot A024 - A096
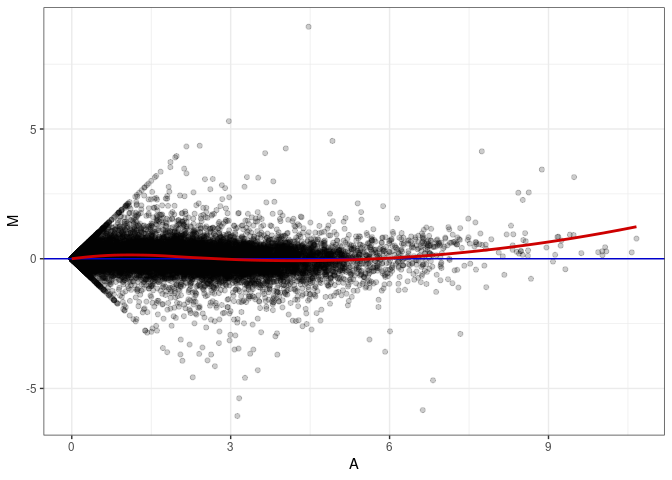
MA plot A024 - A177
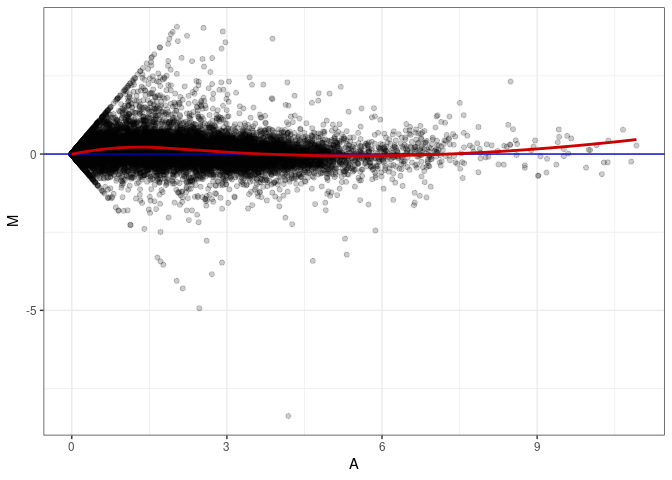
MA plot A033 - A024
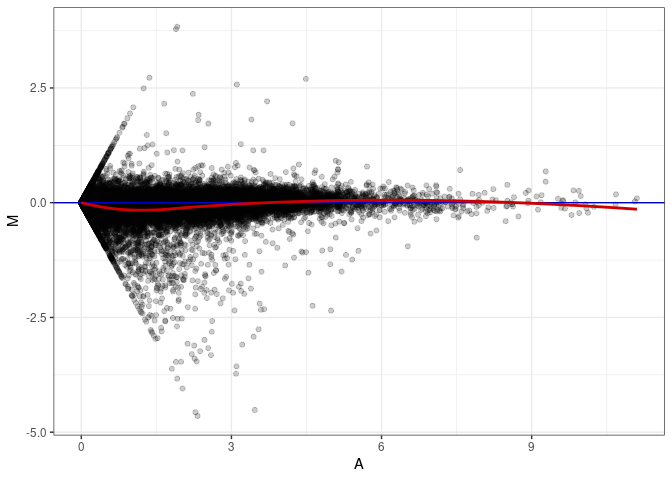
MA plot A033 - A033
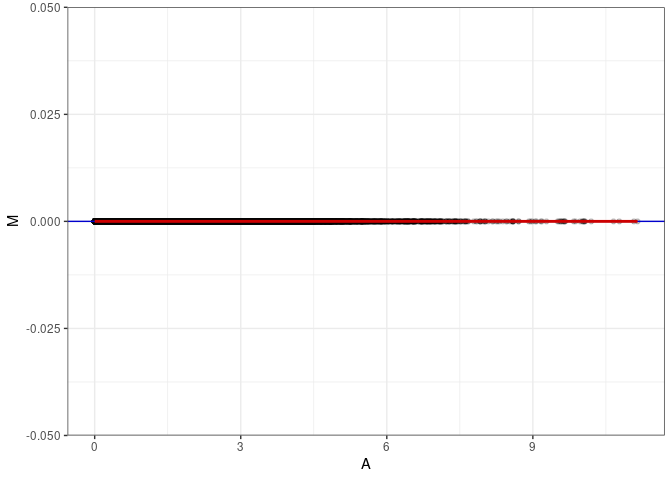
MA plot A033 - A096
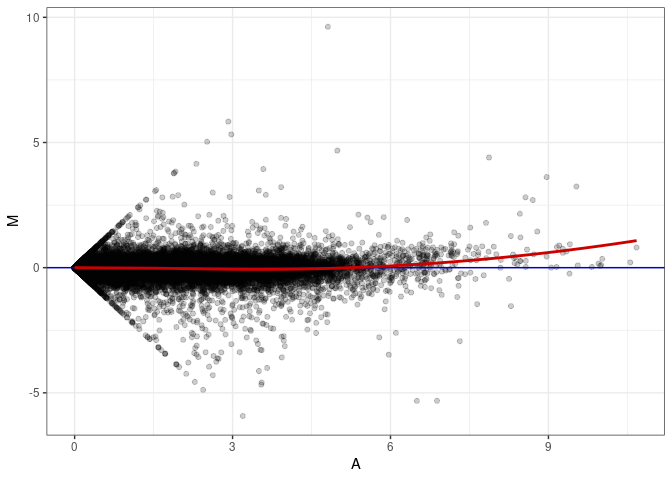
MA plot A033 - A177
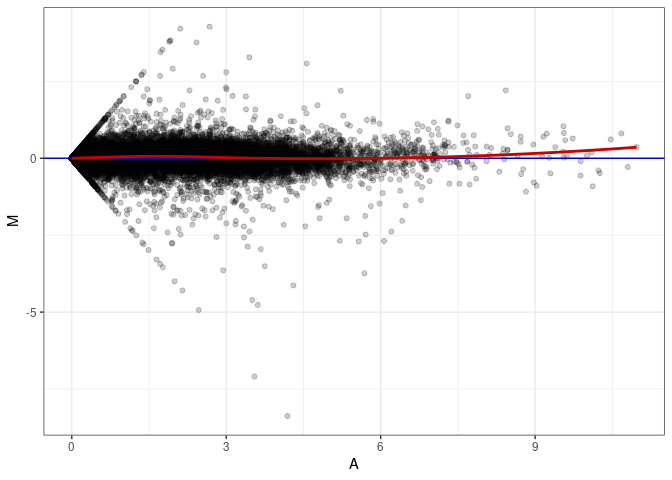
MA plot A096 - A024
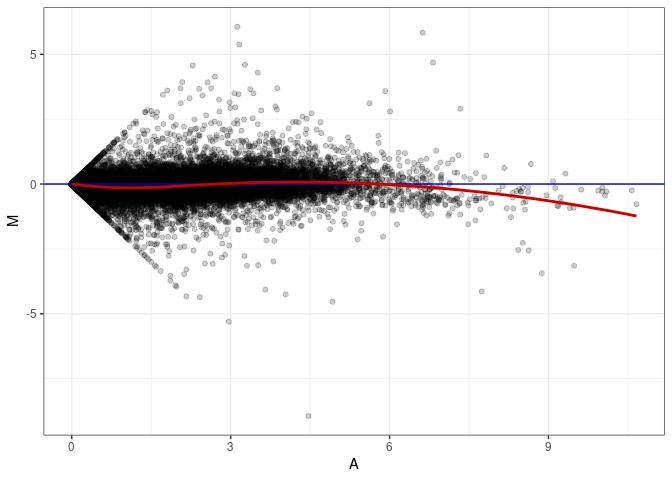
MA plot A096 - A033
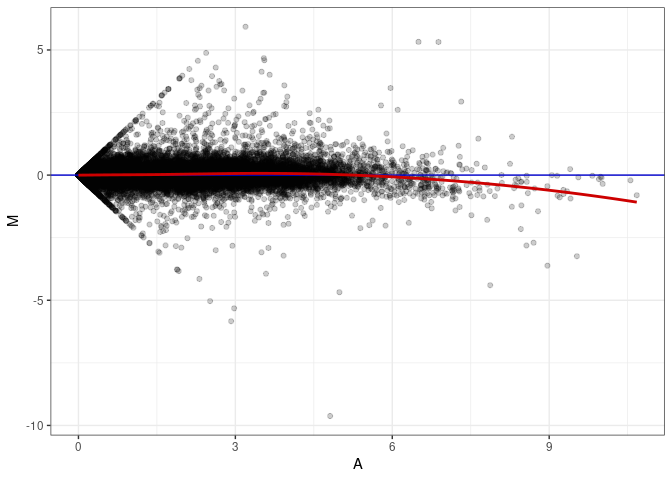
MA plot A096 - A096
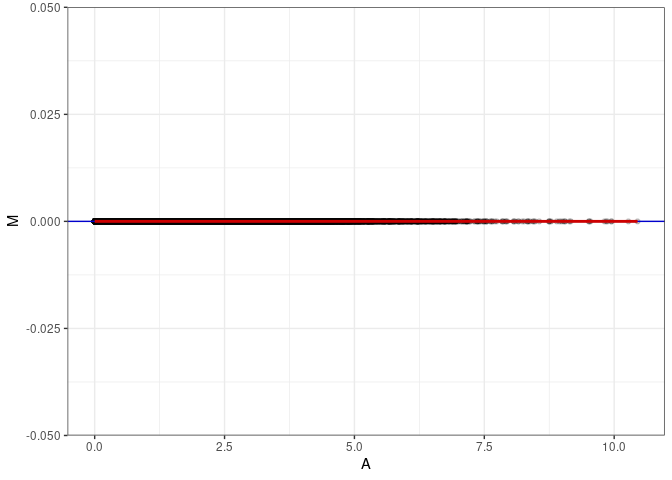
MA plot A096 - A177
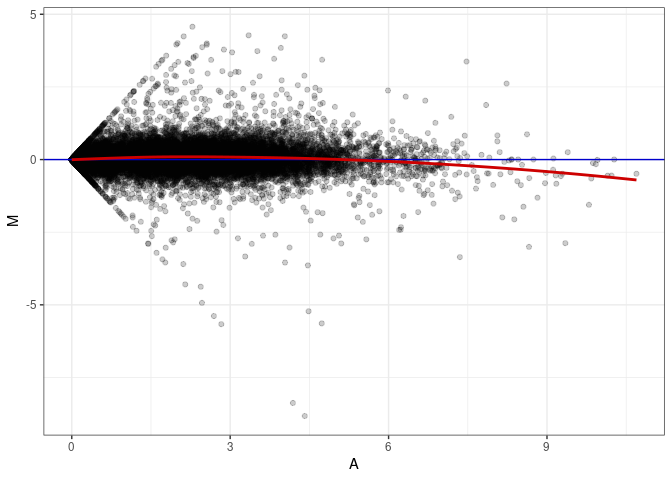
MA plot A177 - A024
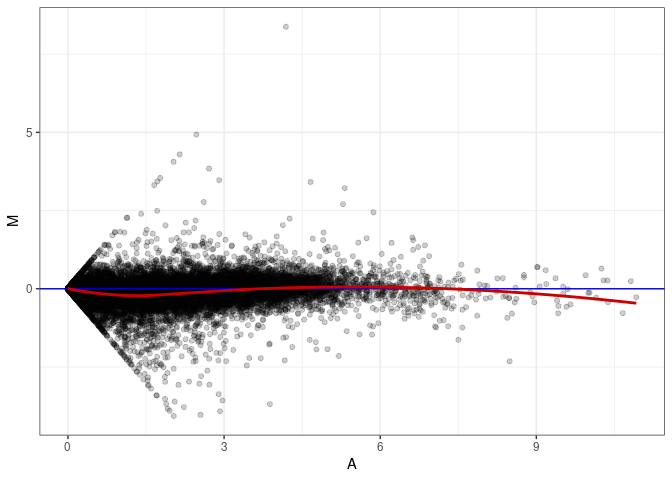
MA plot A177 - A033
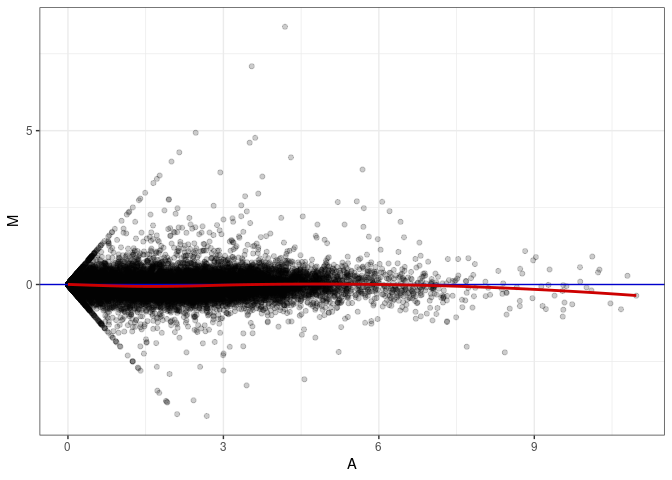
MA plot A177 - A096
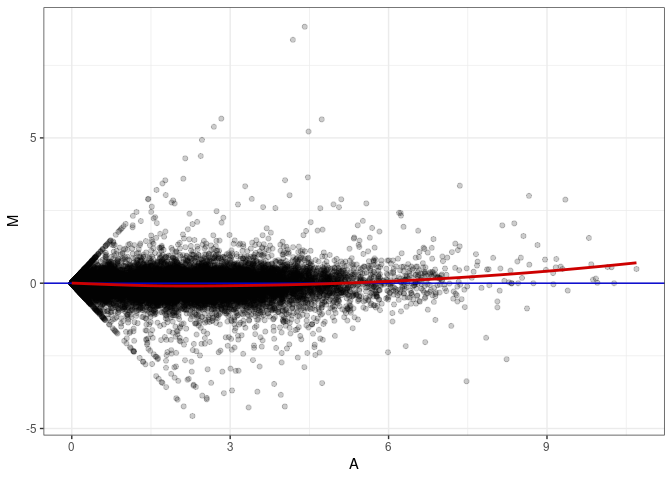
MA plot A177 - A177
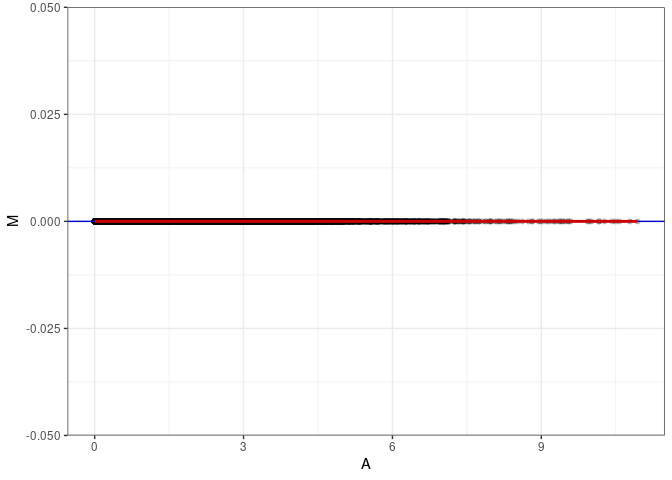
MA plot for Small Intestine control 4wks
subdata <- log.tdata.FPKM.sample.info %>% rownames_to_column() %>% filter(Tissue =="Small Intestine" & Treatment == "None" & Time == "4 wks") %>% column_to_rownames()
subdata <- subdata[,-c(27237:27239)]
log.data.FPKM.sample.info <- t(subdata)
for(i in 1:ncol(log.data.FPKM.sample.info)){
for(j in 1:ncol(log.data.FPKM.sample.info)){
x = log.data.FPKM.sample.info[,i]
y = log.data.FPKM.sample.info[,j]
## M-values
M=x-y ## A-values
A = (x + y)/2
df = data.frame(A, M)
cat('###',"MA plot", colnames(log.data.FPKM.sample.info)[[i]], "-", colnames(log.data.FPKM.sample.info)[[j]] ,' \n')
p <- ggplot(df, aes(x = A, y = M)) + geom_point(size = 1.5, alpha = 1/5) +
geom_hline(color = "blue3", yintercept = 0) + stat_smooth(se = FALSE, method = "loess", color = "red3") + theme_bw()
print(p)
#print(htmltools::tagList(ggplotly(p)))
cat("\n \n")
}
}MA plot A006 - A006
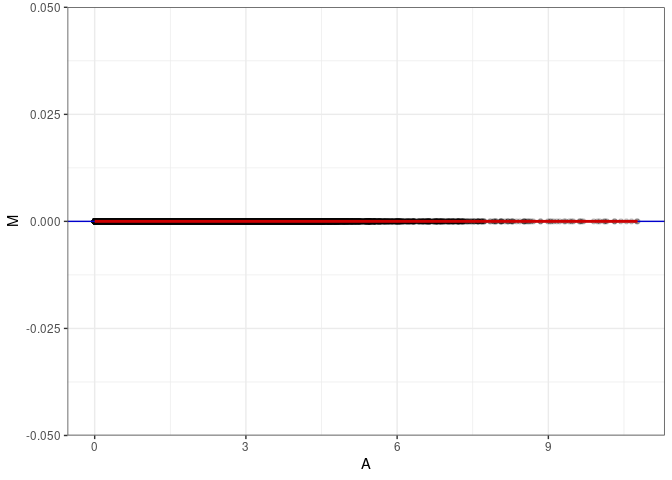
MA plot A006 - A042
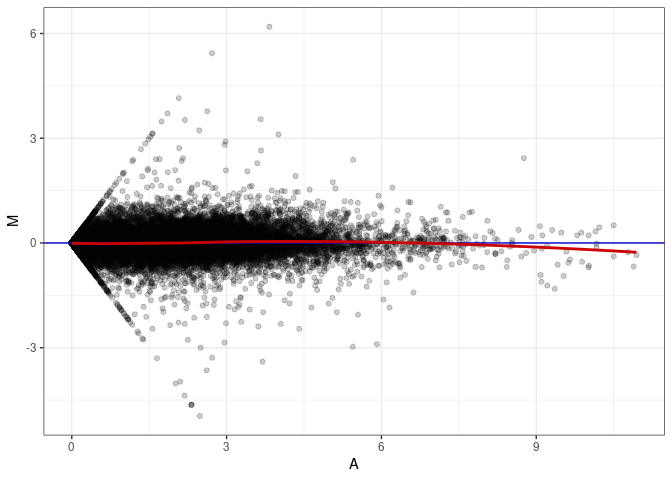
MA plot A006 - A051
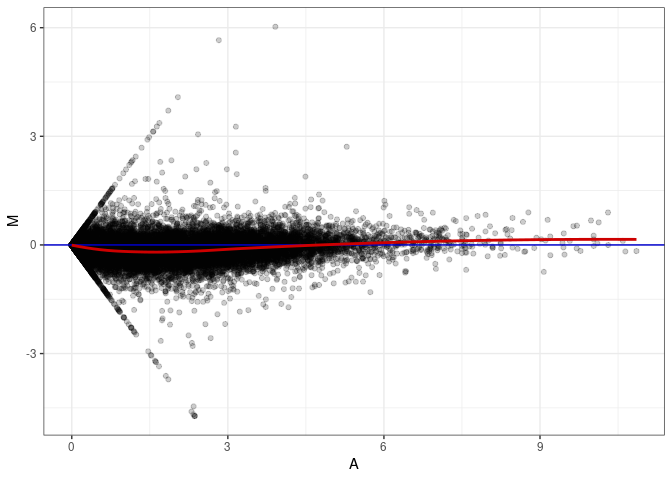
MA plot A006 - A087
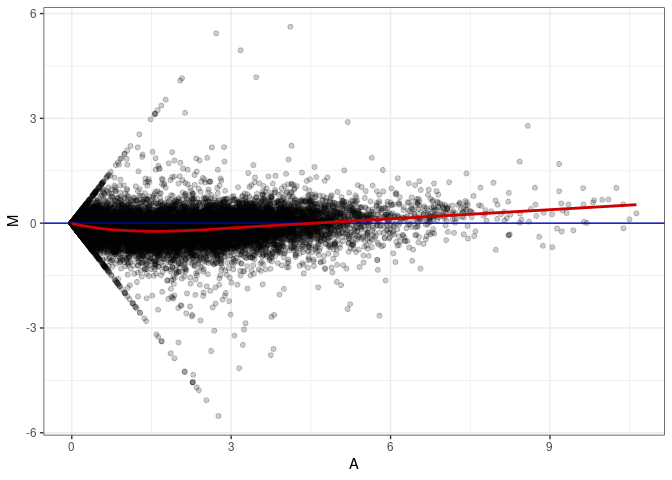
MA plot A042 - A006
MA plot A042 - A042
MA plot A042 - A051
MA plot A042 - A087
MA plot A051 - A006
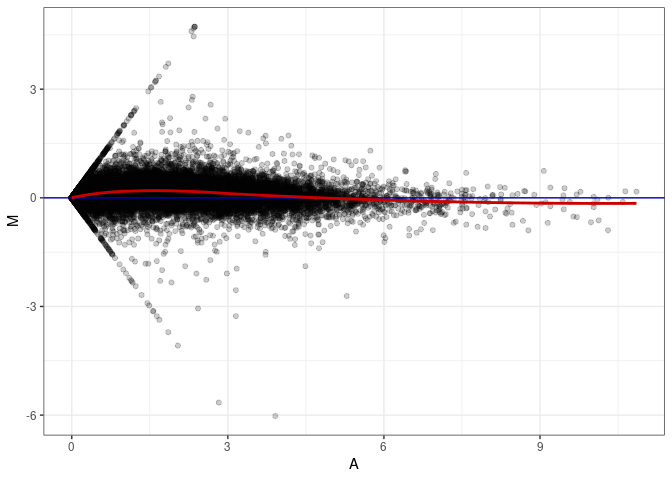
MA plot A051 - A042
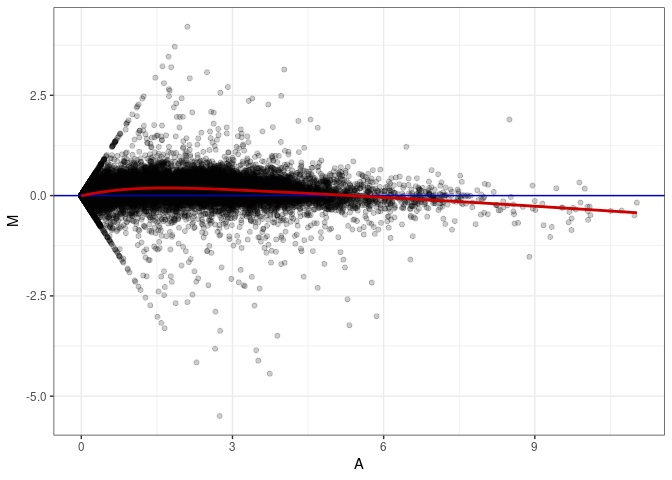
MA plot A051 - A051
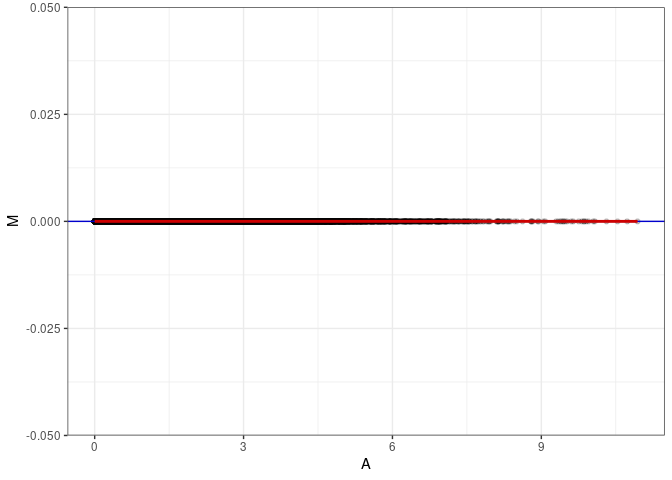
MA plot A051 - A087
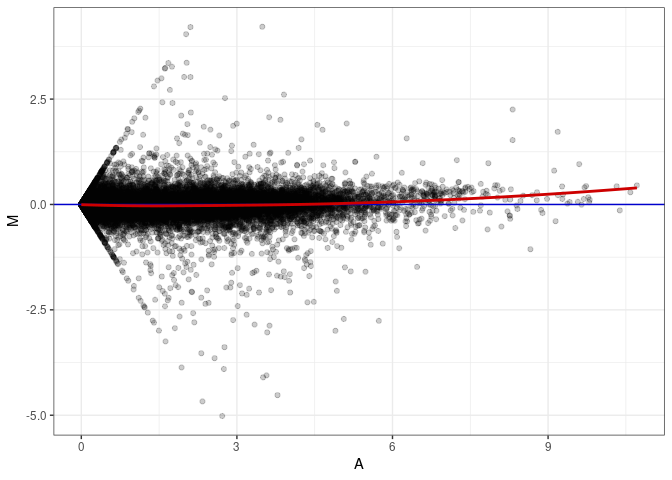
MA plot A087 - A006
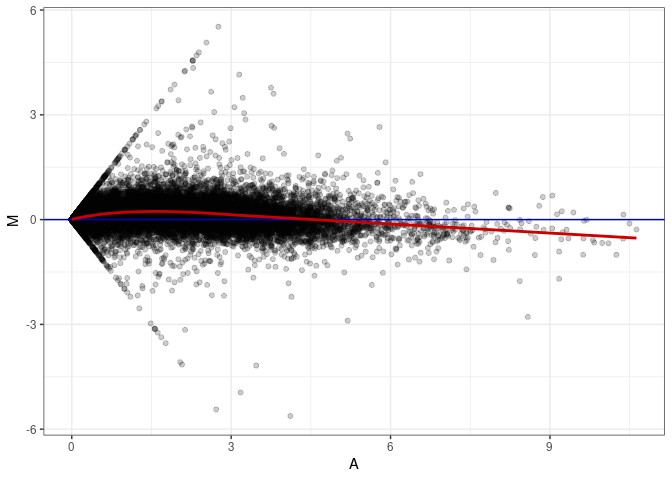
MA plot A087 - A042
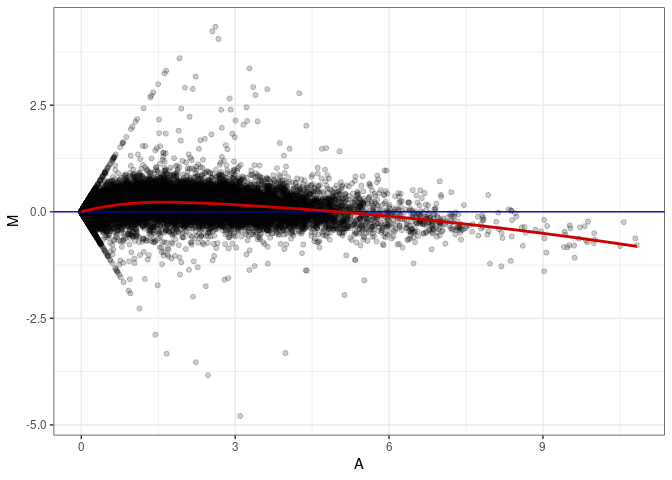
MA plot A087 - A051
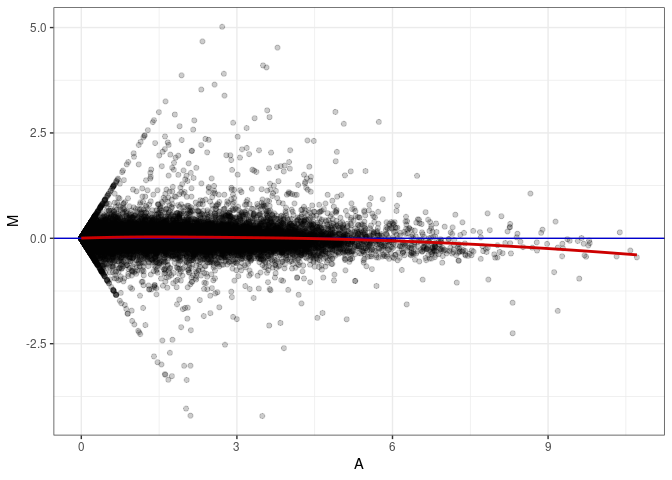
MA plot A087 - A087
MA plot for Small Intestine 2DG 96hrs
subdata <- log.tdata.FPKM.sample.info %>% rownames_to_column() %>% filter(Tissue =="Small Intestine" & Treatment == "2DG" & Time == "96 hrs") %>% column_to_rownames()
subdata <- subdata[,-c(27237:27239)]
log.data.FPKM.sample.info <- t(subdata)
for(i in 1:ncol(log.data.FPKM.sample.info)){
for(j in 1:ncol(log.data.FPKM.sample.info)){
x = log.data.FPKM.sample.info[,i]
y = log.data.FPKM.sample.info[,j]
## M-values
M=x-y ## A-values
A = (x + y)/2
df = data.frame(A, M)
cat('###',"MA plot", colnames(log.data.FPKM.sample.info)[[i]], "-", colnames(log.data.FPKM.sample.info)[[j]] ,' \n')
p <- ggplot(df, aes(x = A, y = M)) + geom_point(size = 1.5, alpha = 1/5) +
geom_hline(color = "blue3", yintercept = 0) + stat_smooth(se = FALSE, method = "loess", color = "red3") + theme_bw()
print(p)
#print(htmltools::tagList(ggplotly(p)))
cat("\n \n")
}
}MA plot A015 - A015
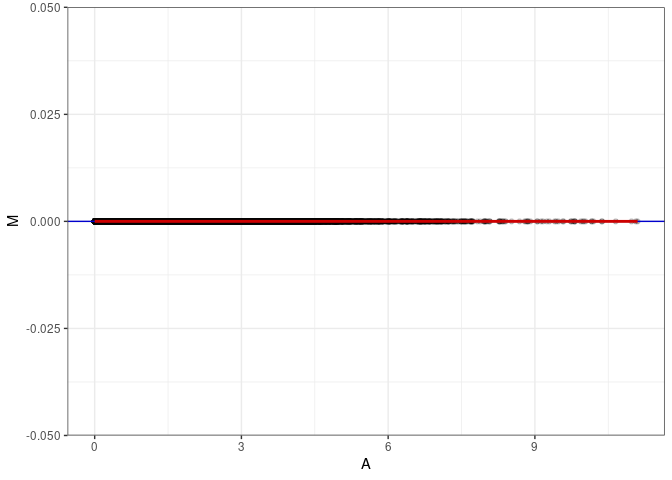
MA plot A015 - A141
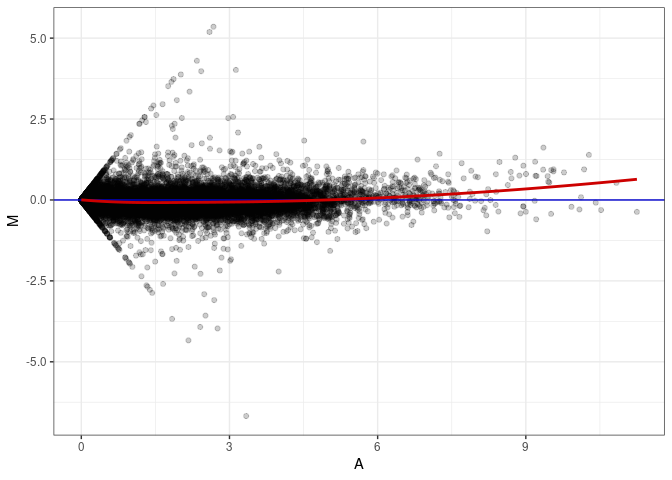
MA plot A015 - A150
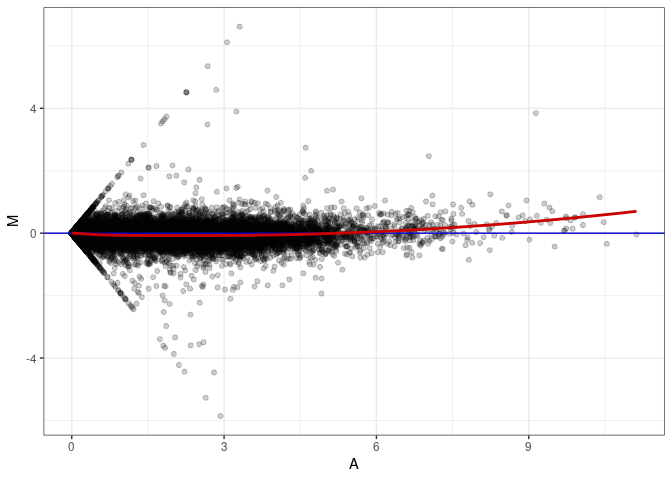
MA plot A015 - A168
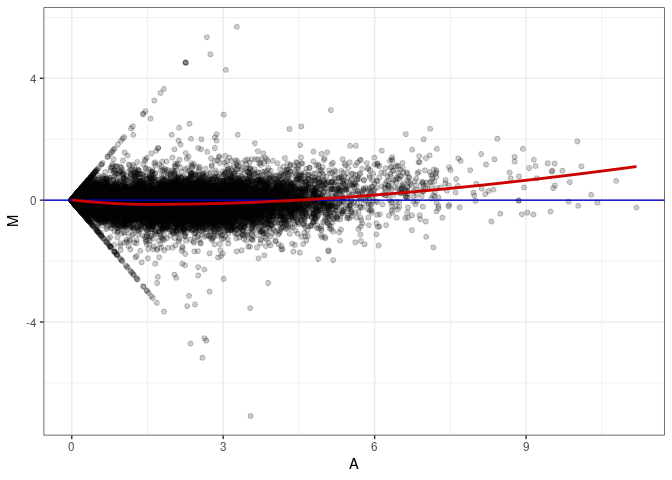
MA plot A141 - A015
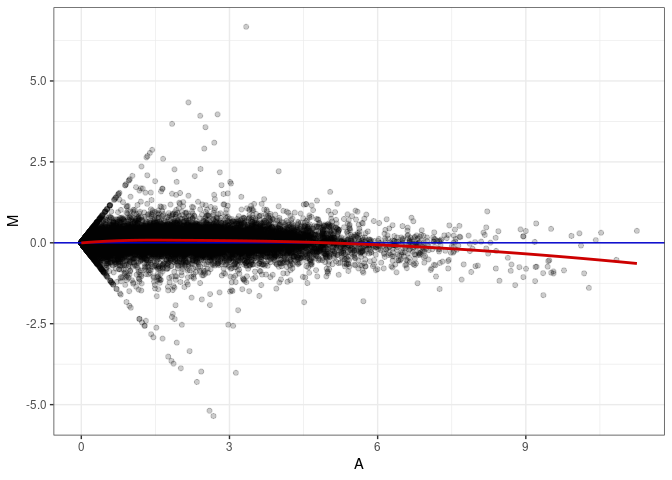
MA plot A141 - A141
MA plot A141 - A150
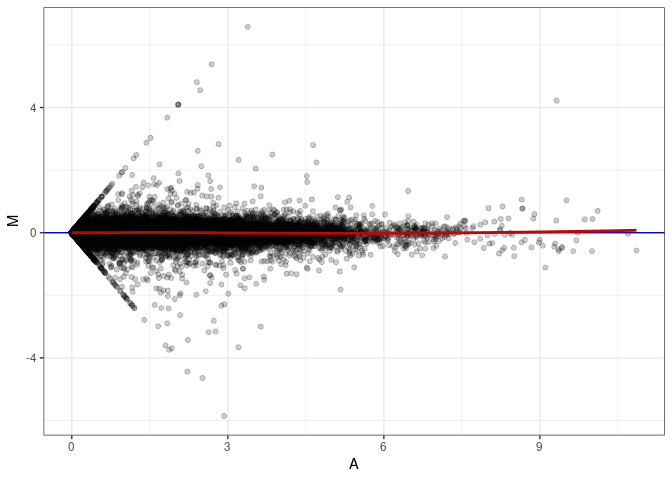
MA plot A141 - A168
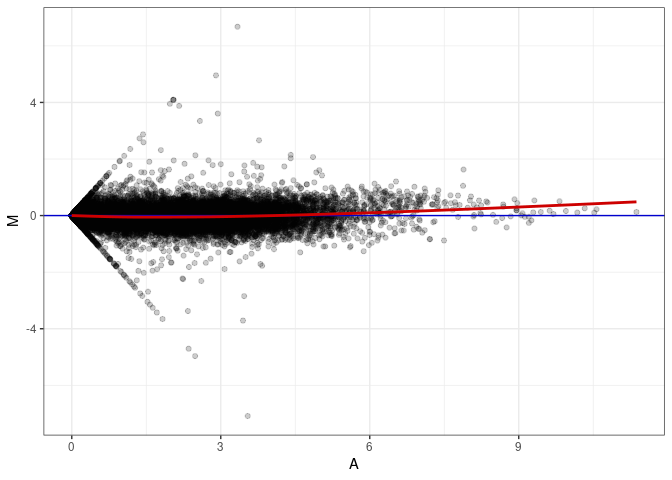
MA plot A150 - A015
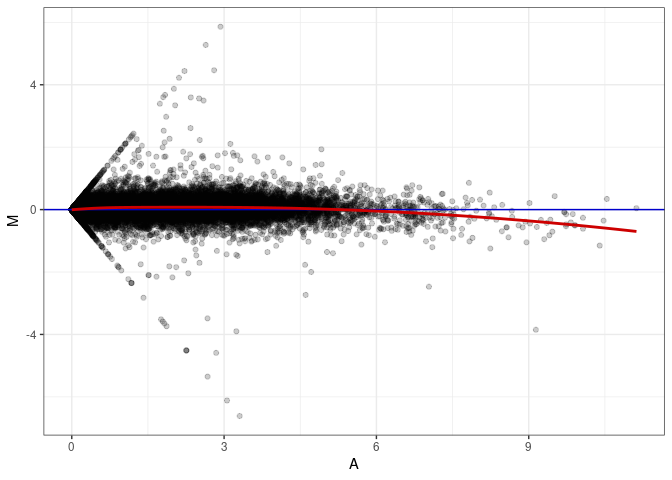
MA plot A150 - A141
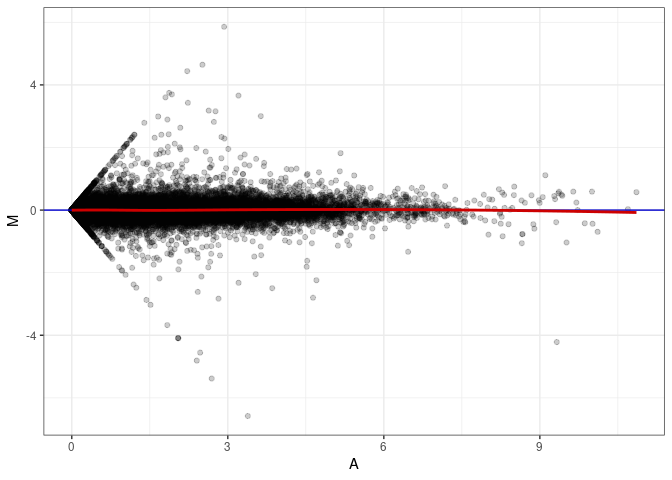
MA plot A150 - A150
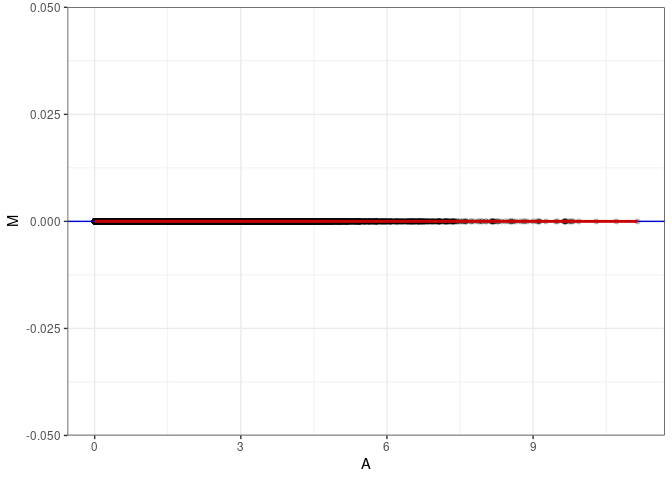
MA plot A150 - A168
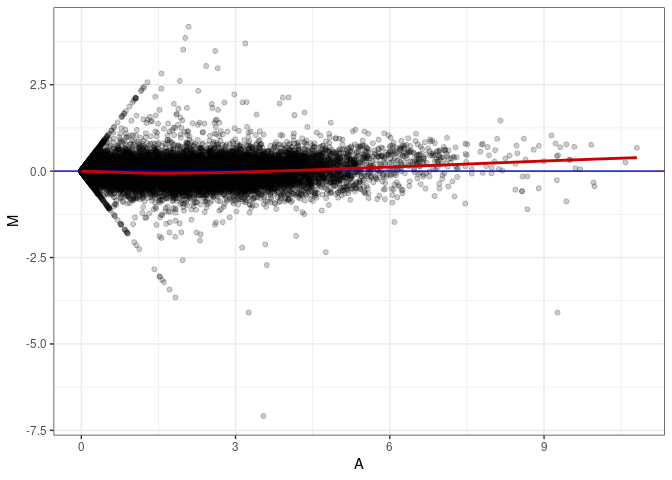
MA plot A168 - A015
MA plot A168 - A141
MA plot A168 - A150
MA plot A168 - A168
MA plot for Small Intestine control 96hrs
subdata <- log.tdata.FPKM.sample.info %>% rownames_to_column() %>% filter(Tissue =="Small Intestine" & Treatment == "None" & Time == "96 hrs") %>% column_to_rownames()
subdata <- subdata[,-c(27237:27239)]
log.data.FPKM.sample.info <- t(subdata)
for(i in 1:ncol(log.data.FPKM.sample.info)){
for(j in 1:ncol(log.data.FPKM.sample.info)){
x = log.data.FPKM.sample.info[,i]
y = log.data.FPKM.sample.info[,j]
## M-values
M=x-y ## A-values
A = (x + y)/2
df = data.frame(A, M)
cat('###',"MA plot", colnames(log.data.FPKM.sample.info)[[i]], "-", colnames(log.data.FPKM.sample.info)[[j]] ,' \n')
p <- ggplot(df, aes(x = A, y = M)) + geom_point(size = 1.5, alpha = 1/5) +
geom_hline(color = "blue3", yintercept = 0) + stat_smooth(se = FALSE, method = "loess", color = "red3") + theme_bw()
print(p)
#print(htmltools::tagList(ggplotly(p)))
cat("\n \n")
}
}MA plot A060 - A060
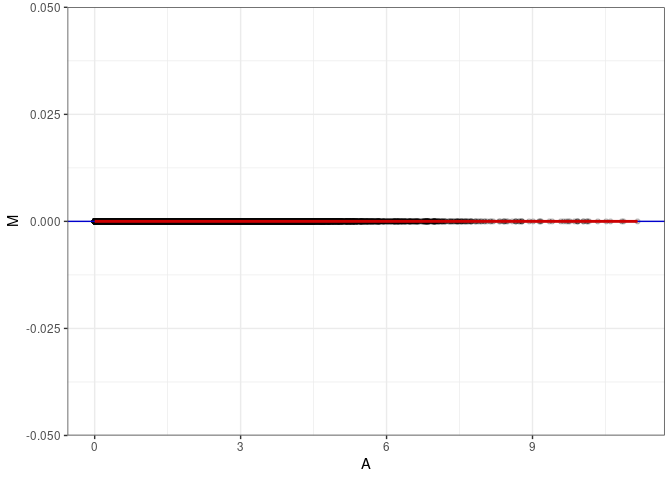
MA plot A060 - A069
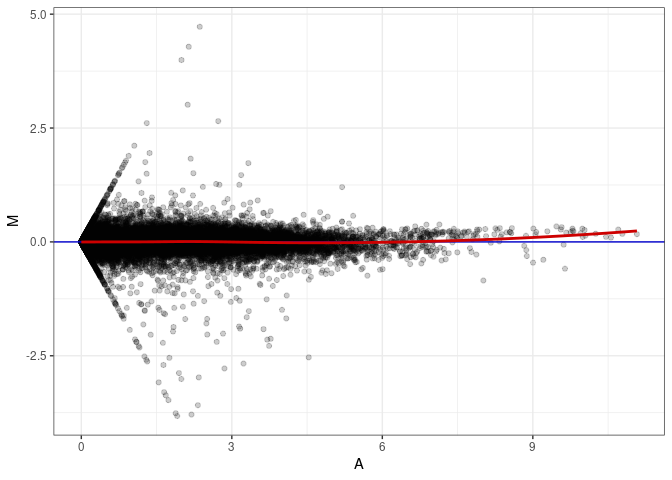
MA plot A060 - A078
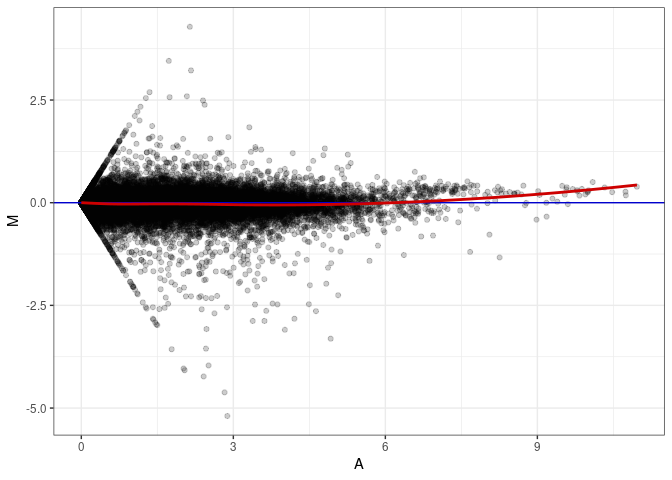
MA plot A060 - A114
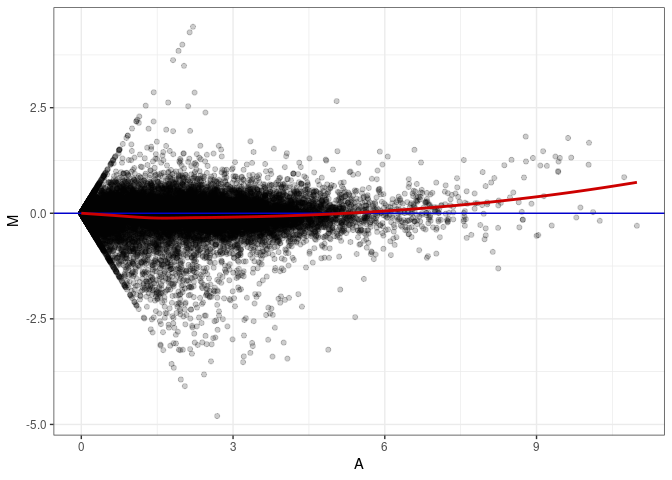
MA plot A069 - A060
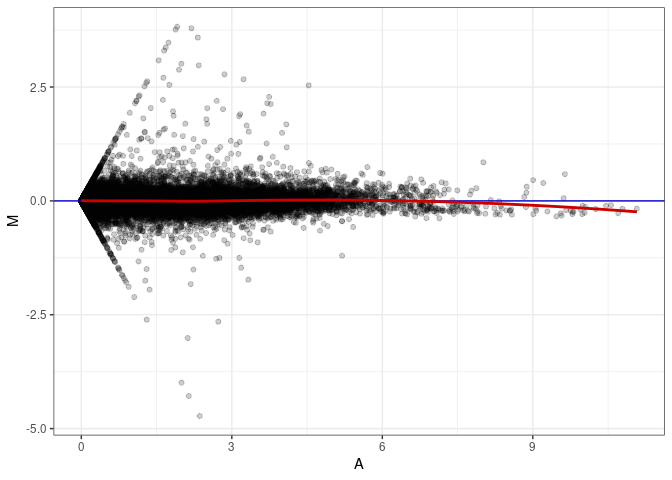
MA plot A069 - A069
MA plot A069 - A078
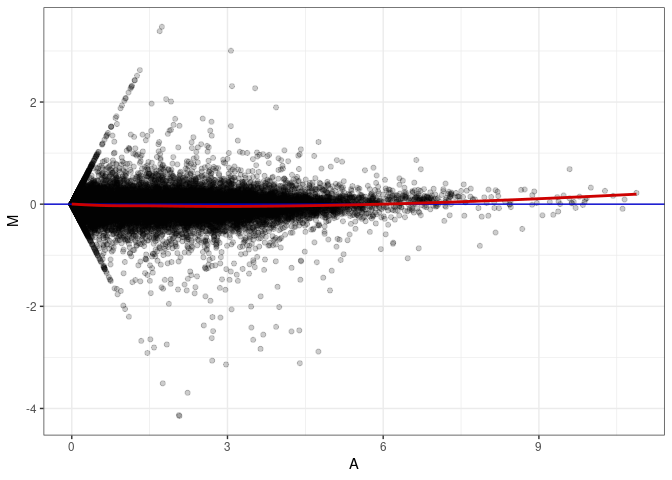
MA plot A069 - A114
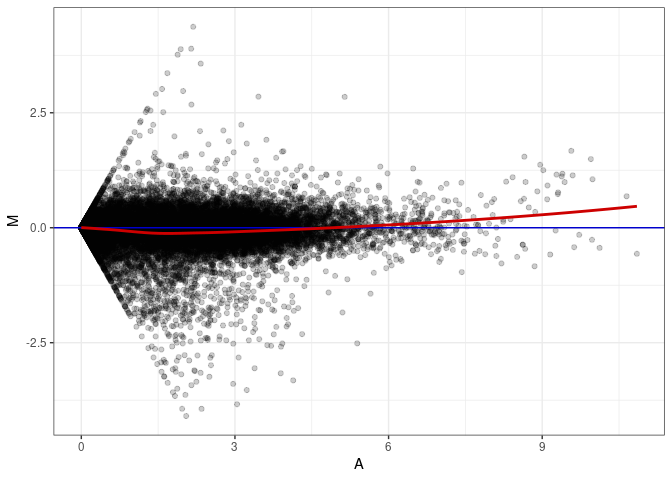
MA plot A078 - A060
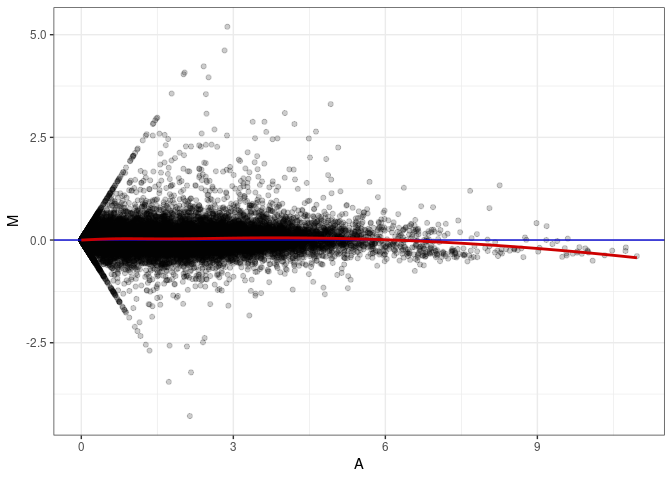
MA plot A078 - A069
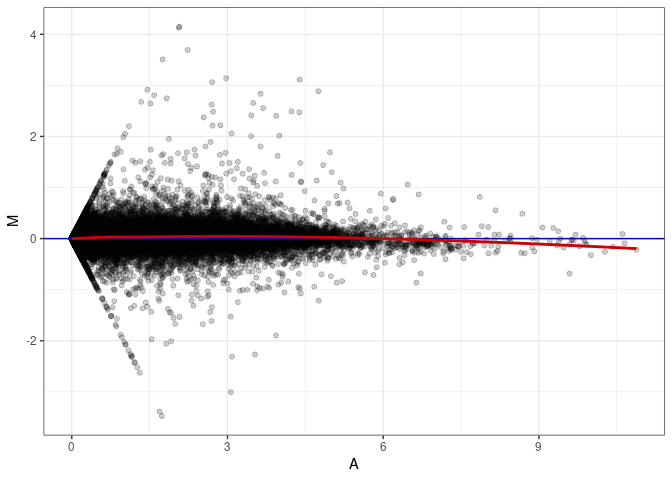
MA plot A078 - A078
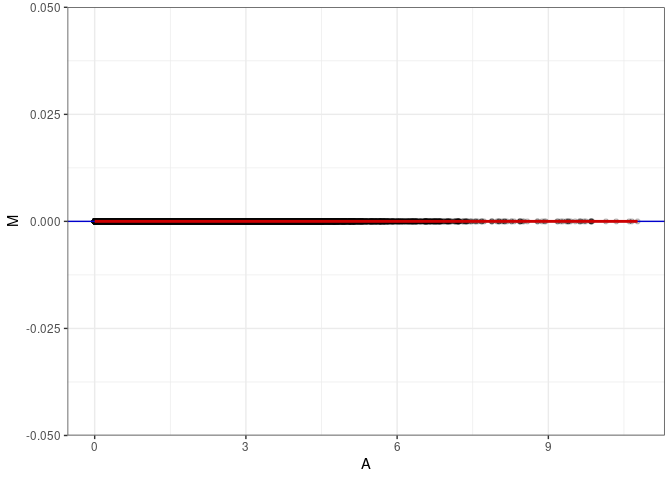
MA plot A078 - A114
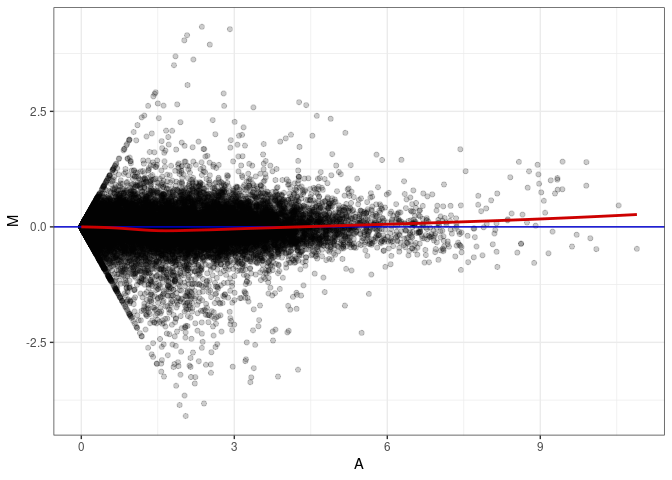
MA plot A114 - A060
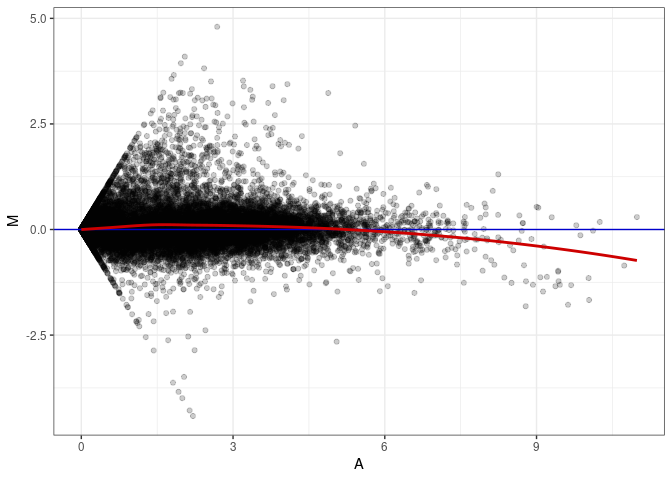
MA plot A114 - A069
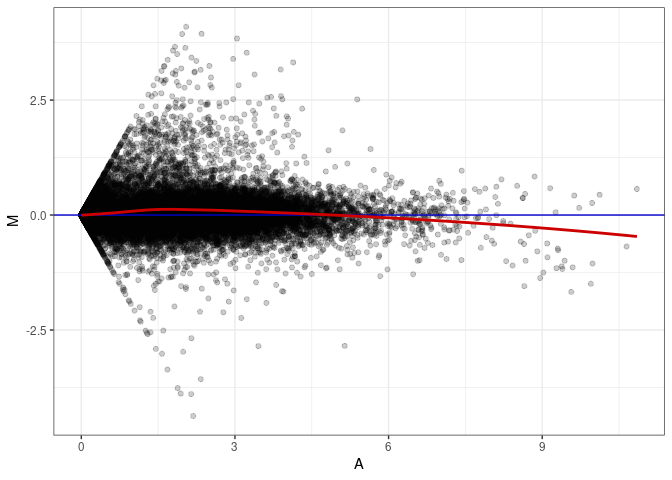
MA plot A114 - A078
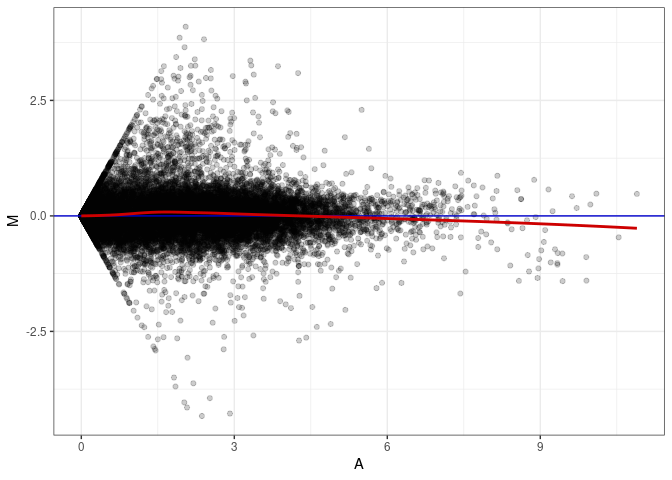
MA plot A114 - A114
MA plot for Skeletal Muscle 2DG 4wks
subdata <- log.tdata.FPKM.sample.info %>% rownames_to_column() %>% filter(Tissue =="Skeletal Muscle" & Treatment == "2DG" & Time == "4 wks") %>% column_to_rownames()
subdata <- subdata[,-c(27237:27239)]
log.data.FPKM.sample.info <- t(subdata)
for(i in 1:ncol(log.data.FPKM.sample.info)){
for(j in 1:ncol(log.data.FPKM.sample.info)){
x = log.data.FPKM.sample.info[,i]
y = log.data.FPKM.sample.info[,j]
## M-values
M=x-y ## A-values
A = (x + y)/2
df = data.frame(A, M)
cat('###',"MA plot", colnames(log.data.FPKM.sample.info)[[i]], "-", colnames(log.data.FPKM.sample.info)[[j]] ,' \n')
p <- ggplot(df, aes(x = A, y = M)) + geom_point(size = 1.5, alpha = 1/5) +
geom_hline(color = "blue3", yintercept = 0) + stat_smooth(se = FALSE, method = "loess", color = "red3") + theme_bw()
print(p)
#print(htmltools::tagList(ggplotly(p)))
cat("\n \n")
}
}MA plot A023 - A023
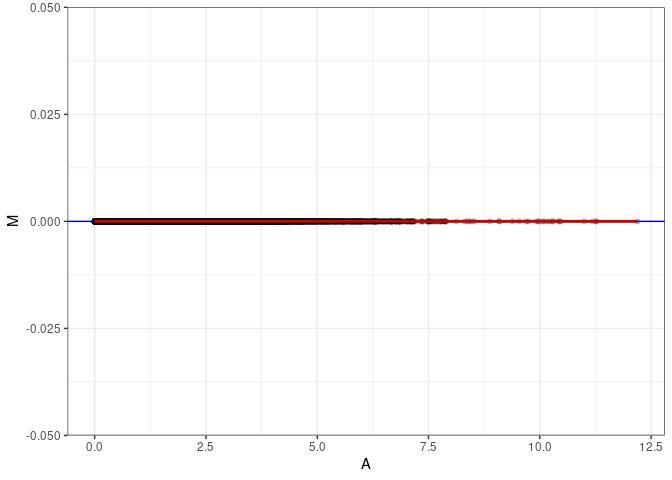
MA plot A023 - A032
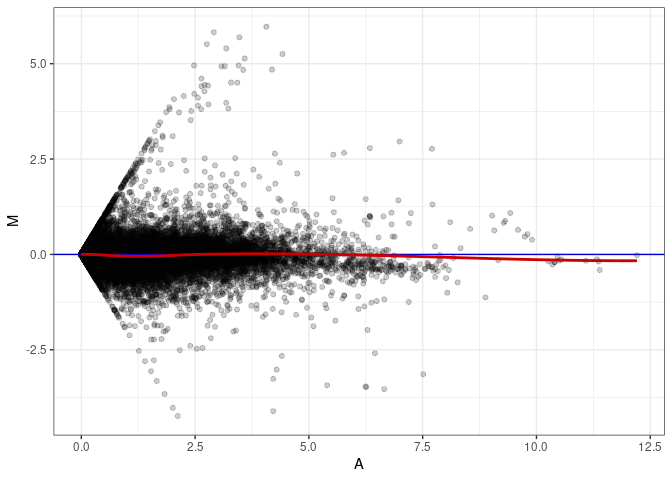
MA plot A023 - A095
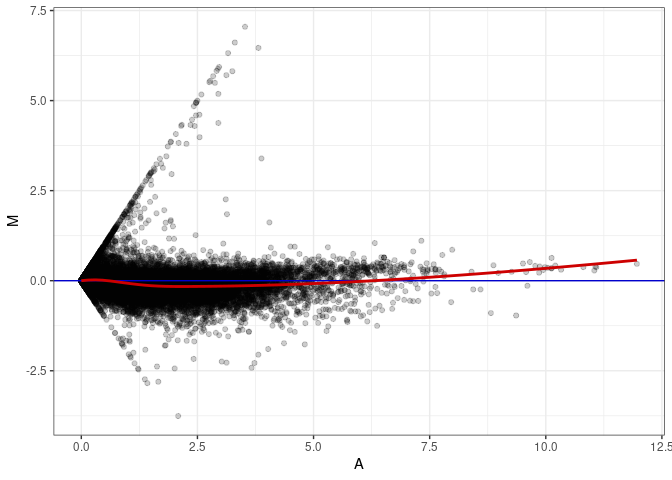
MA plot A023 - A176
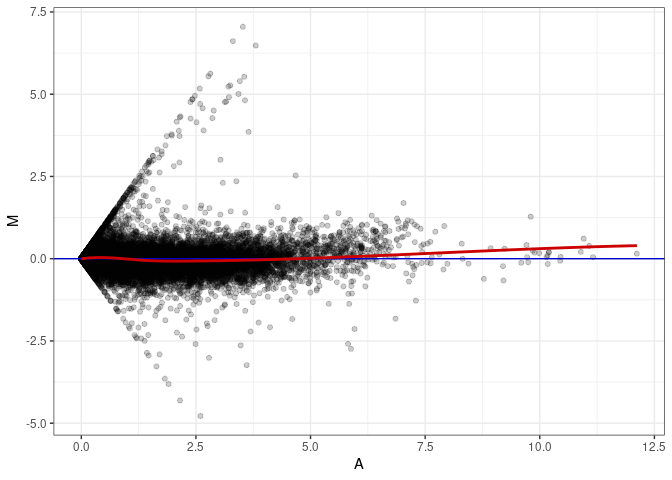
MA plot A032 - A023
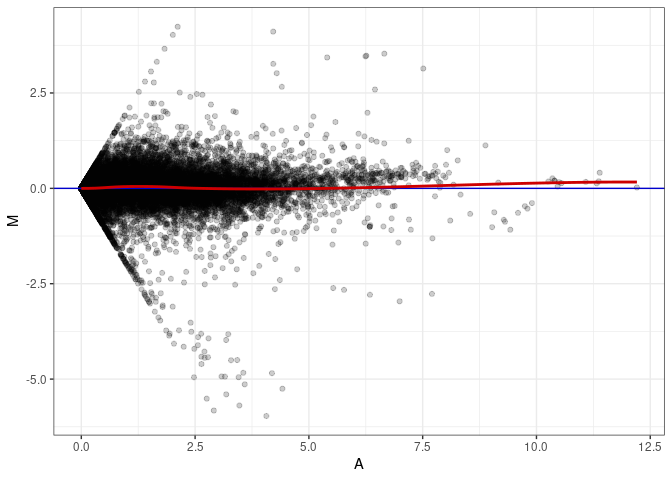
MA plot A032 - A032
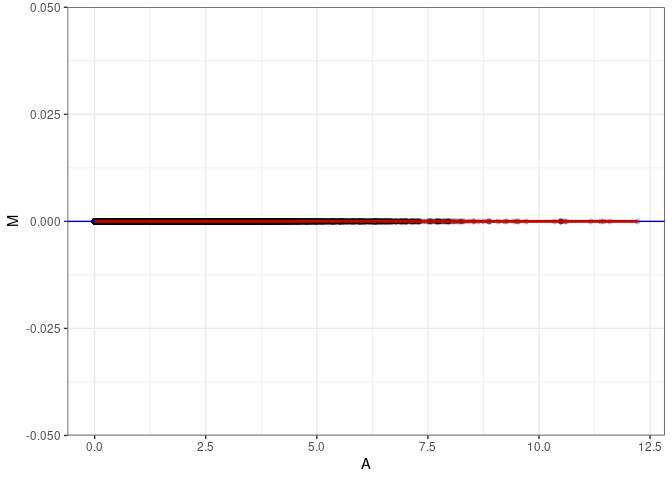
MA plot A032 - A095
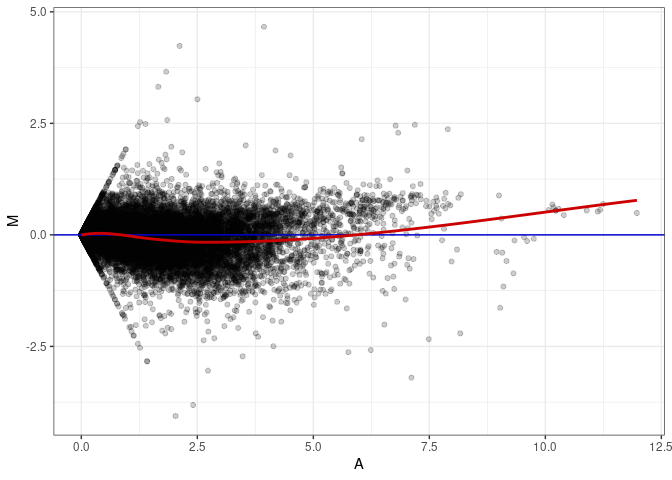
MA plot A032 - A176
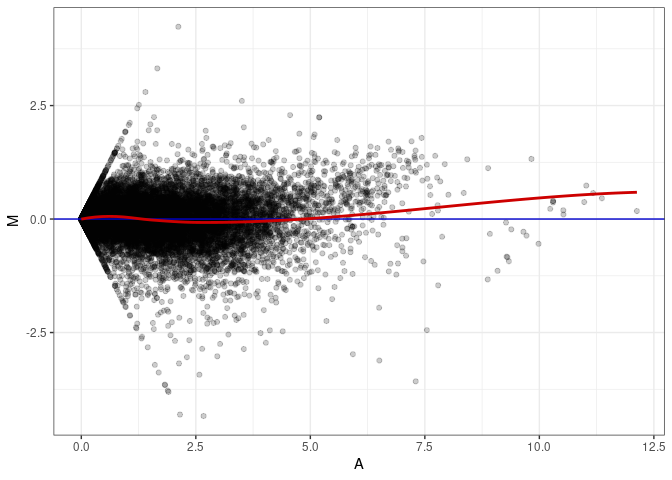
MA plot A095 - A023
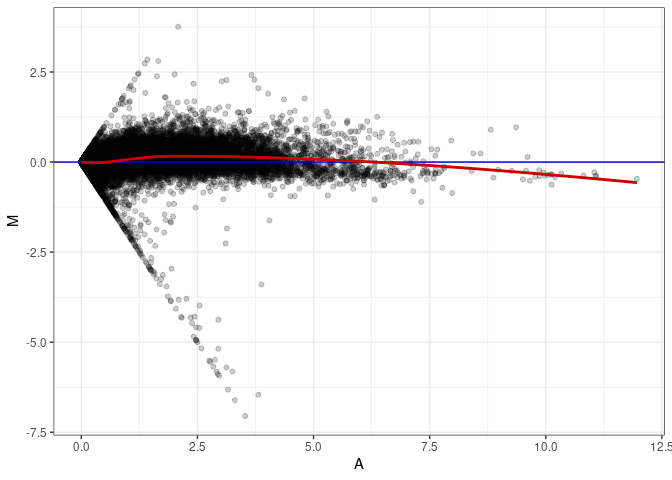
MA plot A095 - A032
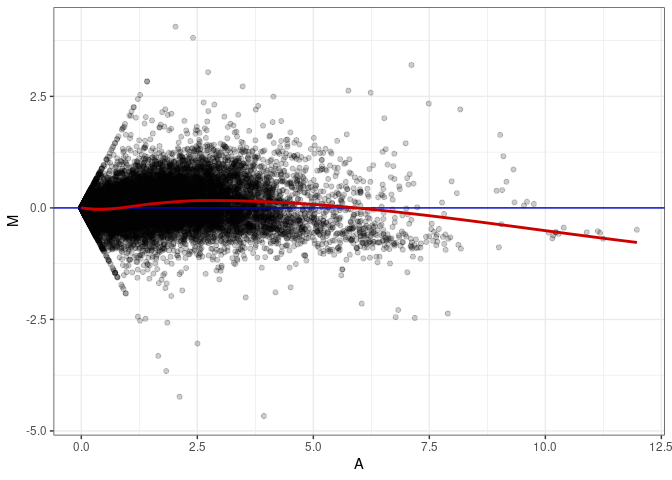
MA plot A095 - A095
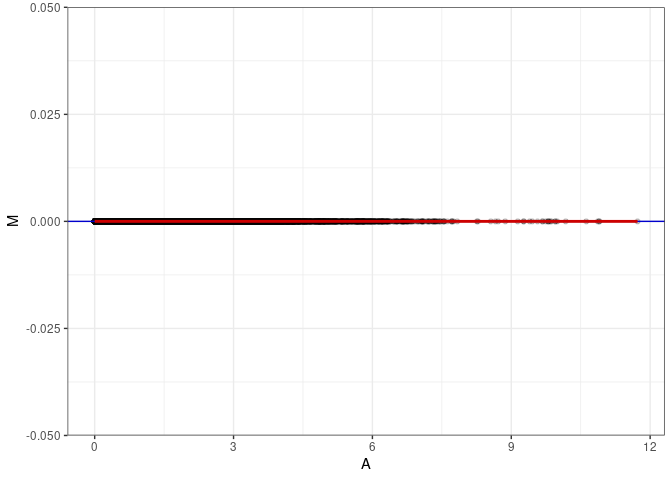
MA plot A095 - A176
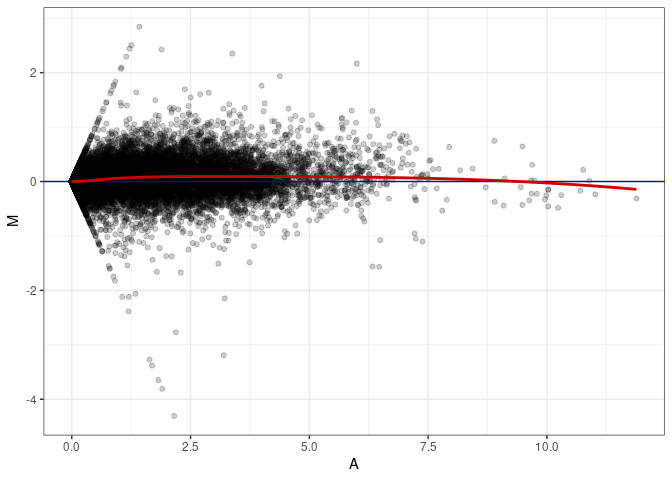
MA plot A176 - A023
MA plot A176 - A032
MA plot A176 - A095
MA plot A176 - A176
MA plot for Skeletal Muscle control 4wks
subdata <- log.tdata.FPKM.sample.info %>% rownames_to_column() %>% filter(Tissue =="Skeletal Muscle" & Treatment == "None" & Time == "4 wks") %>% column_to_rownames()
subdata <- subdata[,-c(27237:27239)]
log.data.FPKM.sample.info <- t(subdata)
for(i in 1:ncol(log.data.FPKM.sample.info)){
for(j in 1:ncol(log.data.FPKM.sample.info)){
x = log.data.FPKM.sample.info[,i]
y = log.data.FPKM.sample.info[,j]
## M-values
M=x-y ## A-values
A = (x + y)/2
df = data.frame(A, M)
cat('###',"MA plot", colnames(log.data.FPKM.sample.info)[[i]], "-", colnames(log.data.FPKM.sample.info)[[j]] ,' \n')
p <- ggplot(df, aes(x = A, y = M)) + geom_point(size = 1.5, alpha = 1/5) +
geom_hline(color = "blue3", yintercept = 0) + stat_smooth(se = FALSE, method = "loess", color = "red3") + theme_bw()
print(p)
#print(htmltools::tagList(ggplotly(p)))
cat("\n \n")
}
}MA plot A005 - A005
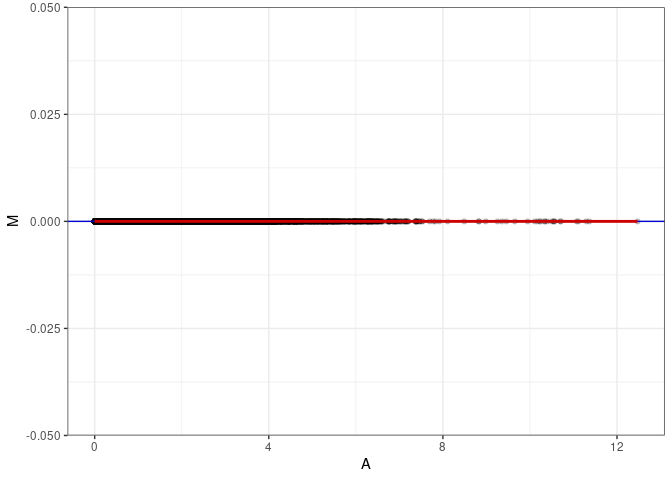
MA plot A005 - A041
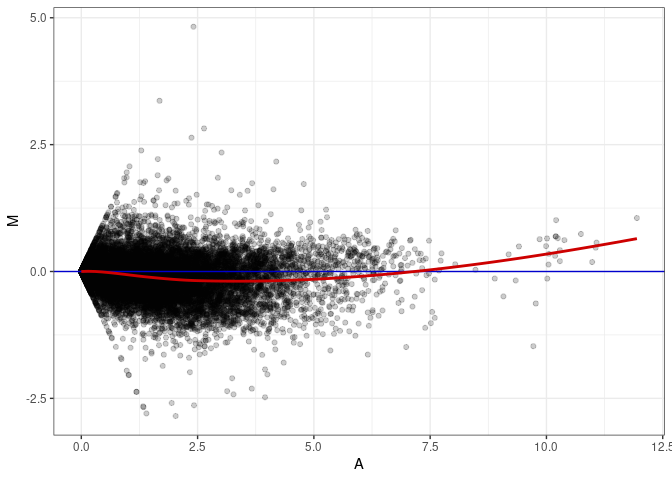
MA plot A005 - A050
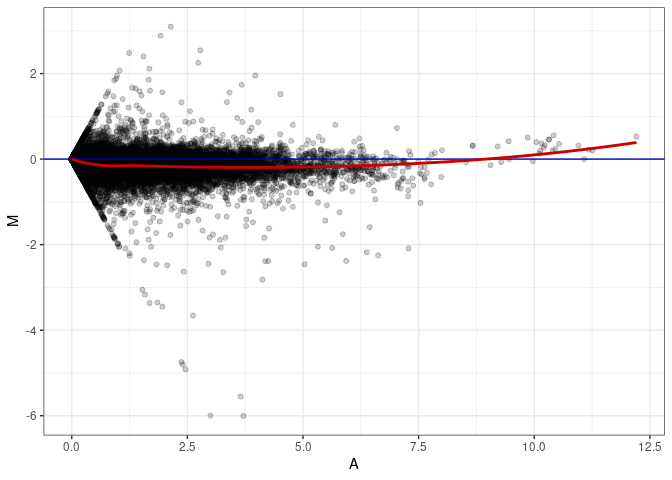
MA plot A005 - A086
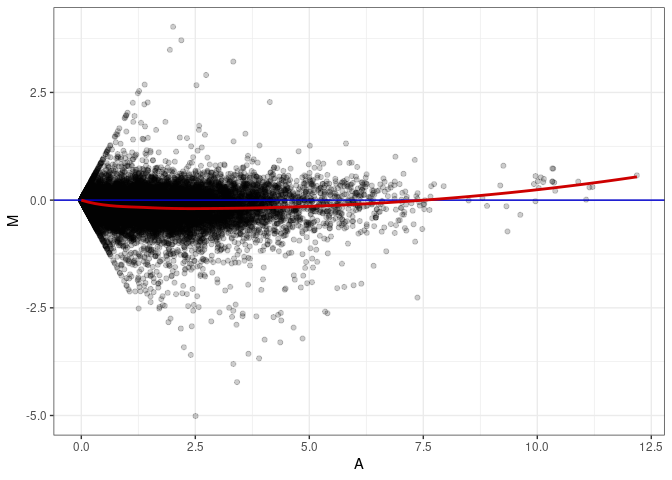
MA plot A041 - A005
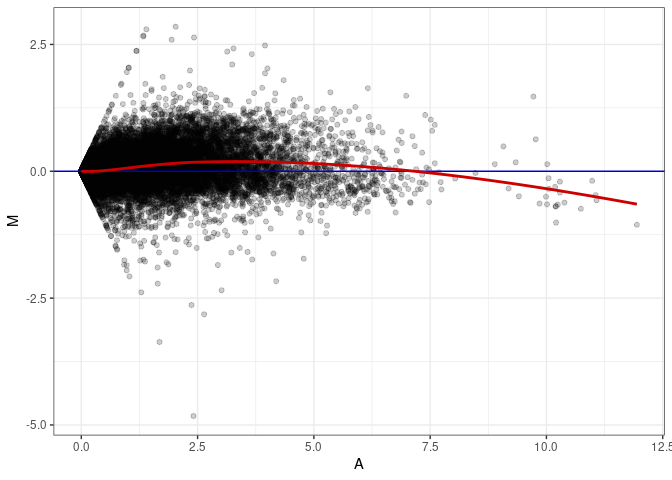
MA plot A041 - A041
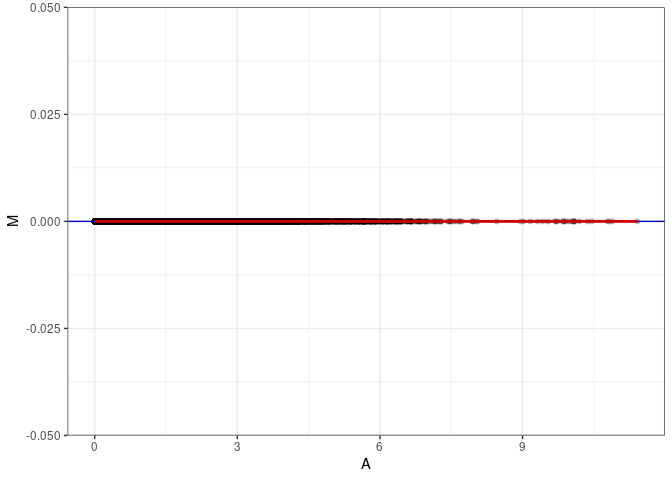
MA plot A041 - A050
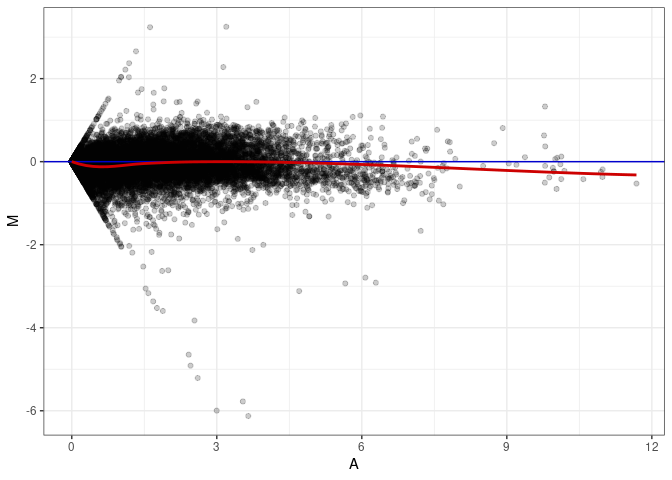
MA plot A041 - A086
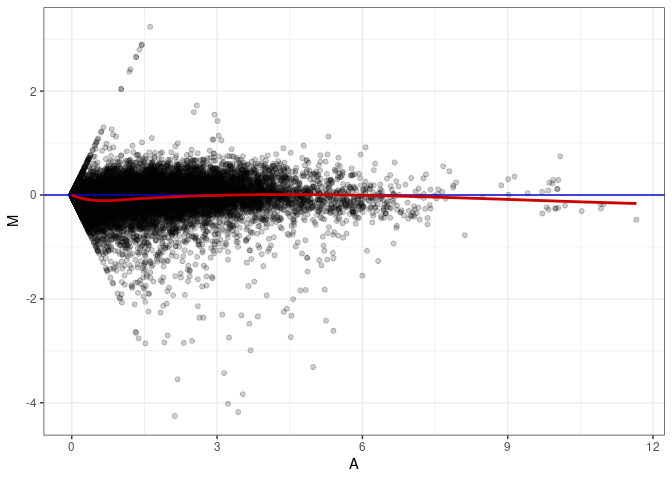
MA plot A050 - A005
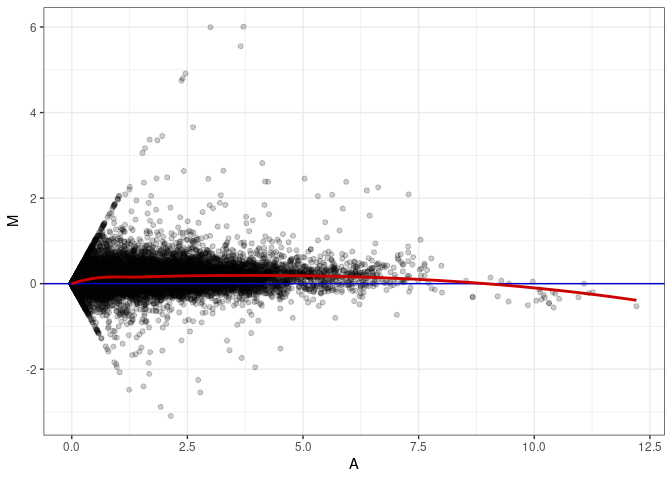
MA plot A050 - A041
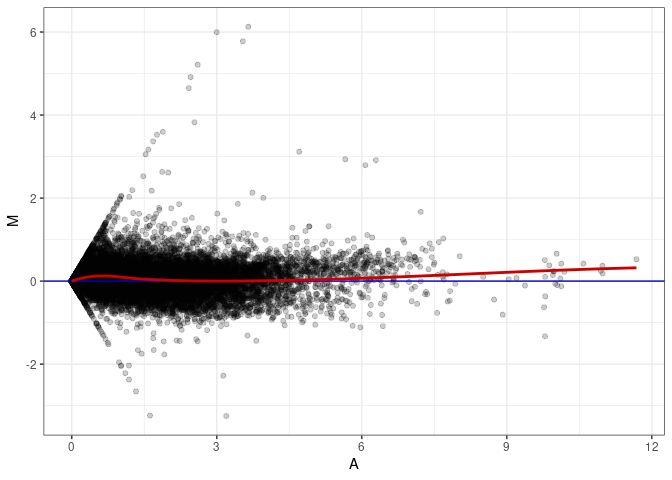
MA plot A050 - A050
MA plot A050 - A086
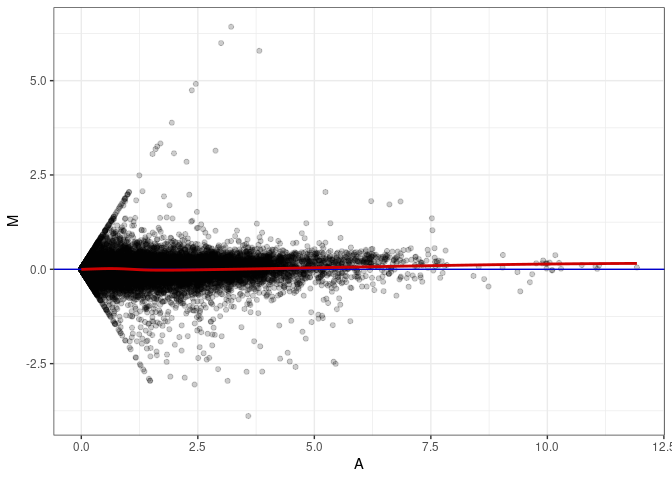
MA plot A086 - A005
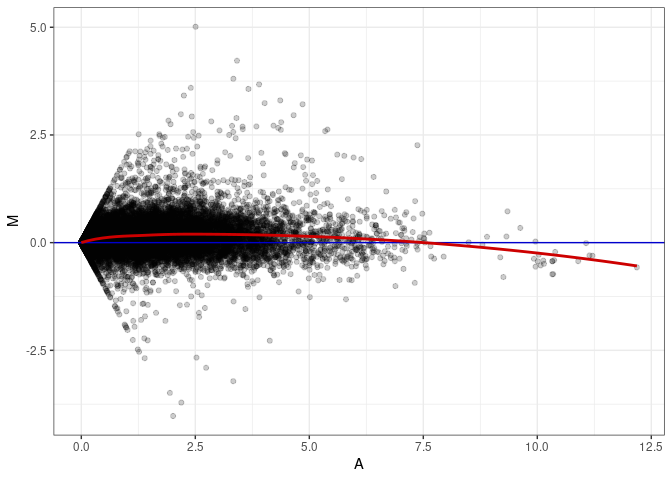
MA plot A086 - A041
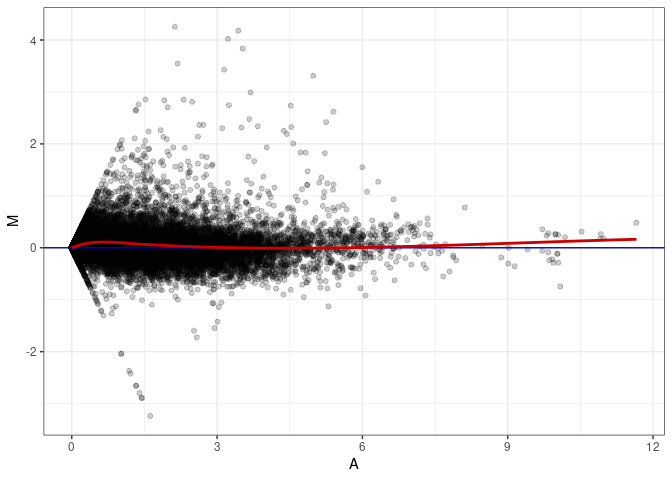
MA plot A086 - A050
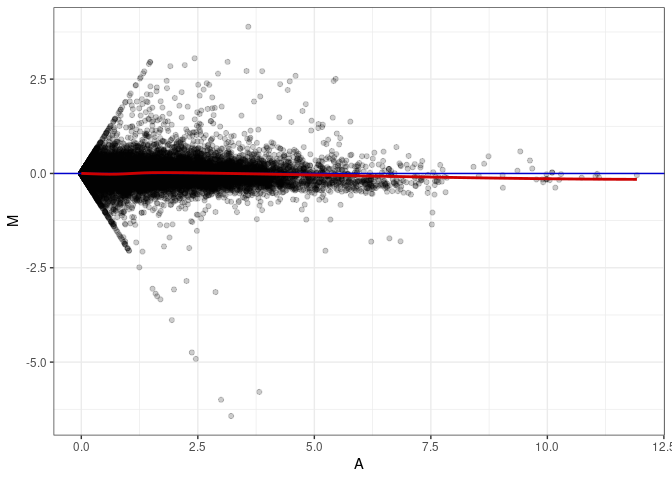
MA plot A086 - A086
MA plot for Skeletal Muscle 2DG 96hrs
subdata <- log.tdata.FPKM.sample.info %>% rownames_to_column() %>% filter(Tissue =="Skeletal Muscle" & Treatment == "2DG" & Time == "96 hrs") %>% column_to_rownames()
subdata <- subdata[,-c(27237:27239)]
log.data.FPKM.sample.info <- t(subdata)
for(i in 1:ncol(log.data.FPKM.sample.info)){
for(j in 1:ncol(log.data.FPKM.sample.info)){
x = log.data.FPKM.sample.info[,i]
y = log.data.FPKM.sample.info[,j]
## M-values
M=x-y ## A-values
A = (x + y)/2
df = data.frame(A, M)
cat('###',"MA plot", colnames(log.data.FPKM.sample.info)[[i]], "-", colnames(log.data.FPKM.sample.info)[[j]] ,' \n')
p <- ggplot(df, aes(x = A, y = M)) + geom_point(size = 1.5, alpha = 1/5) +
geom_hline(color = "blue3", yintercept = 0) + stat_smooth(se = FALSE, method = "loess", color = "red3") + theme_bw()
print(p)
#print(htmltools::tagList(ggplotly(p)))
cat("\n \n")
}
}MA plot A014 - A014
MA plot A014 - A140
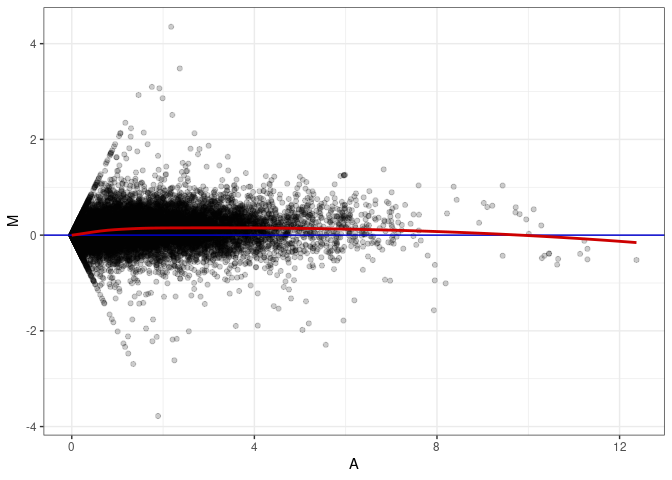
MA plot A014 - A149
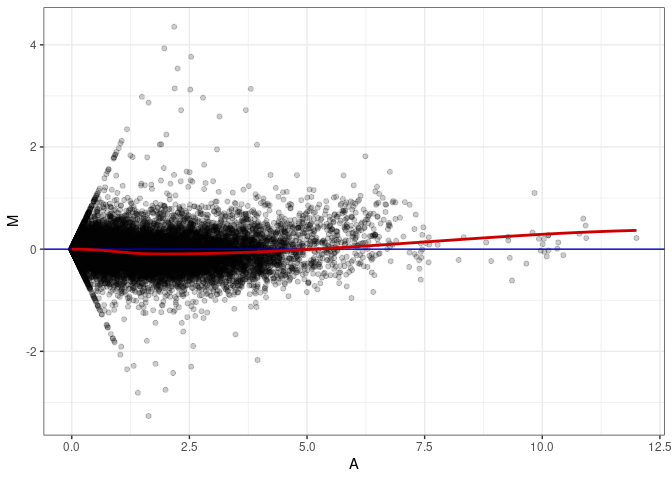
MA plot A014 - A167
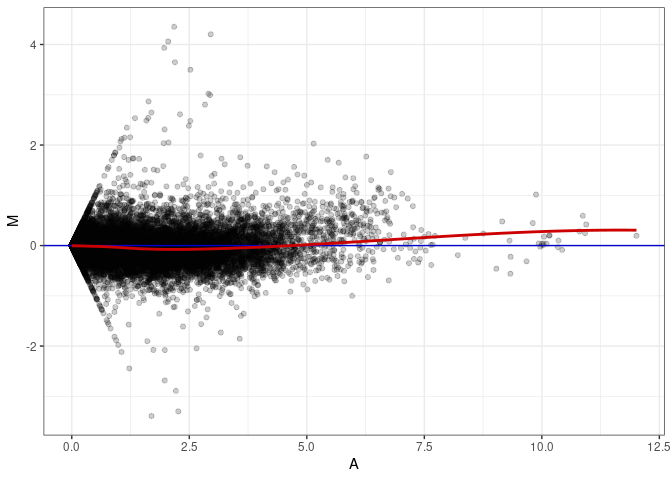
MA plot A140 - A014
MA plot A140 - A140
MA plot A140 - A149
MA plot A140 - A167
MA plot A149 - A014
MA plot A149 - A140
MA plot A149 - A149
MA plot A149 - A167
MA plot A167 - A014
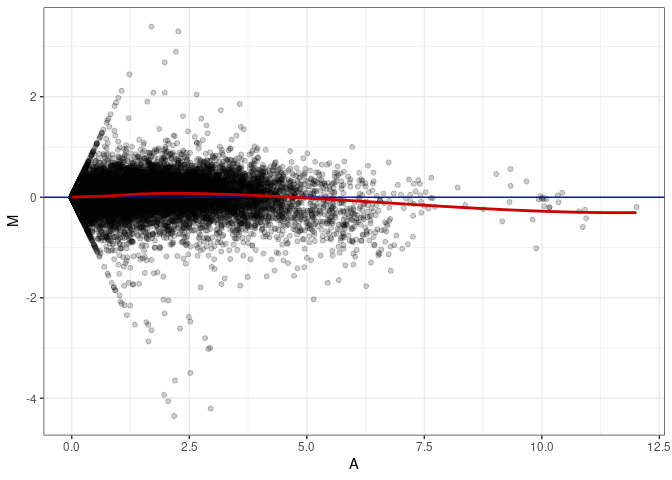
MA plot A167 - A140
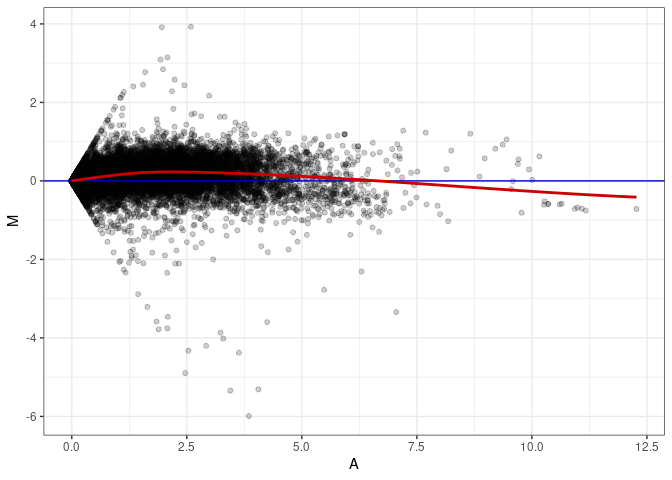
MA plot A167 - A149
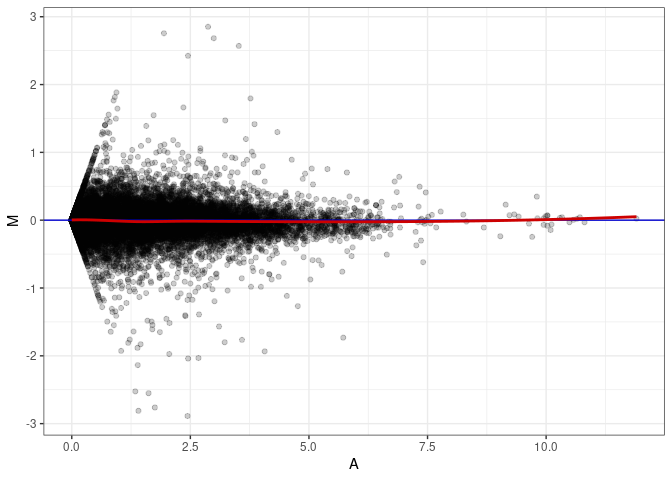
MA plot A167 - A167
MA plot for Skeletal Muscle control 96hrs
subdata <- log.tdata.FPKM.sample.info %>% rownames_to_column() %>% filter(Tissue =="Skeletal Muscle" & Treatment == "None" & Time == "96 hrs") %>% column_to_rownames()
subdata <- subdata[,-c(27237:27239)]
log.data.FPKM.sample.info <- t(subdata)
for(i in 1:ncol(log.data.FPKM.sample.info)){
for(j in 1:ncol(log.data.FPKM.sample.info)){
x = log.data.FPKM.sample.info[,i]
y = log.data.FPKM.sample.info[,j]
## M-values
M=x-y ## A-values
A = (x + y)/2
df = data.frame(A, M)
cat('###',"MA plot", colnames(log.data.FPKM.sample.info)[[i]], "-", colnames(log.data.FPKM.sample.info)[[j]] ,' \n')
p <- ggplot(df, aes(x = A, y = M)) + geom_point(size = 1.5, alpha = 1/5) +
geom_hline(color = "blue3", yintercept = 0) + stat_smooth(se = FALSE, method = "loess", color = "red3") + theme_bw()
print(p)
#print(htmltools::tagList(ggplotly(p)))
cat("\n \n")
}
}MA plot A059 - A059
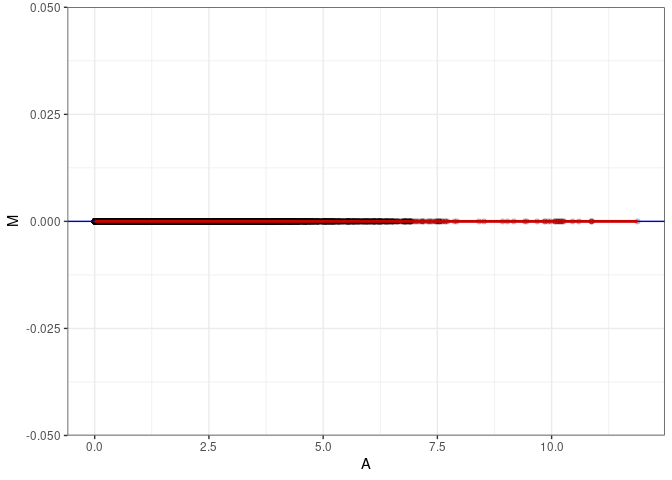
MA plot A059 - A068
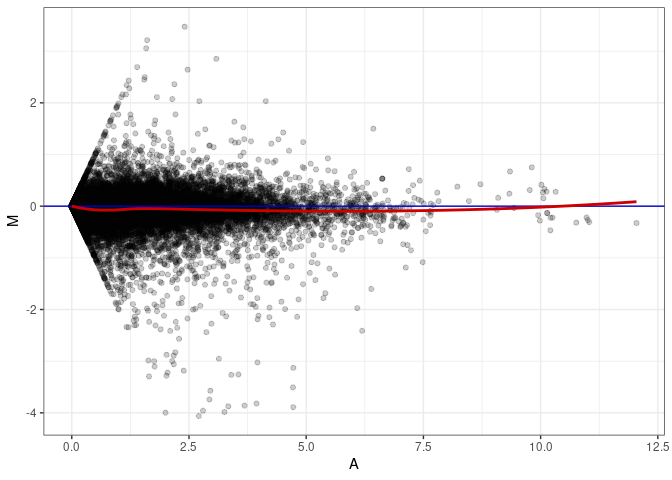
MA plot A059 - A077
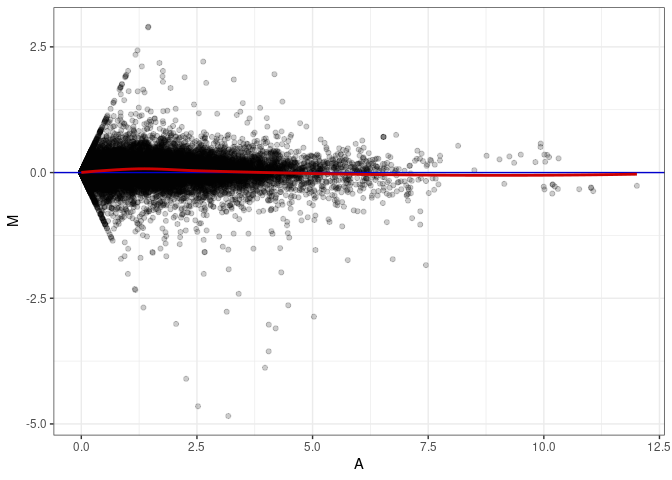
MA plot A059 - A113
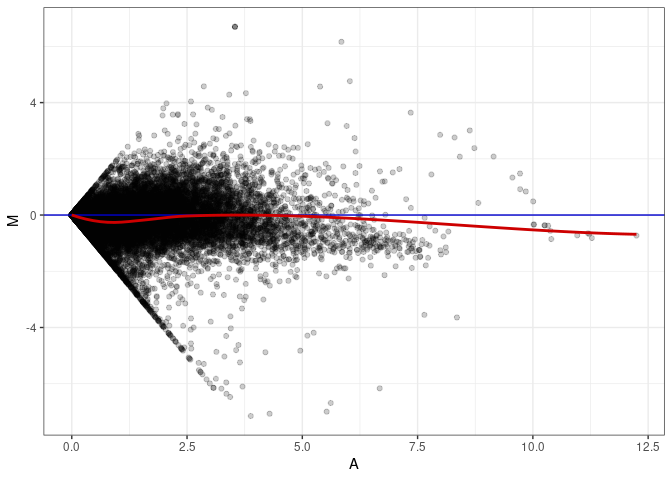
MA plot A068 - A059
MA plot A068 - A068
MA plot A068 - A077
MA plot A068 - A113
MA plot A077 - A059
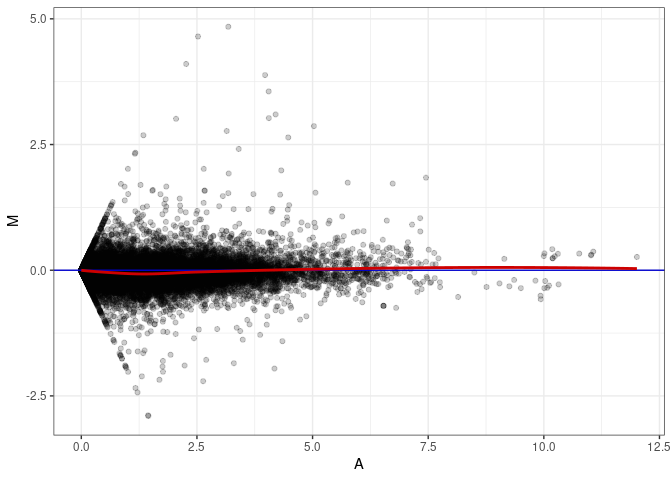
MA plot A077 - A068
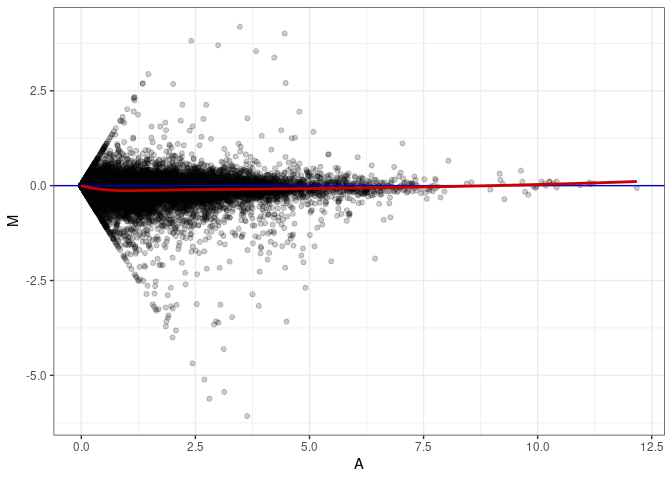
MA plot A077 - A077
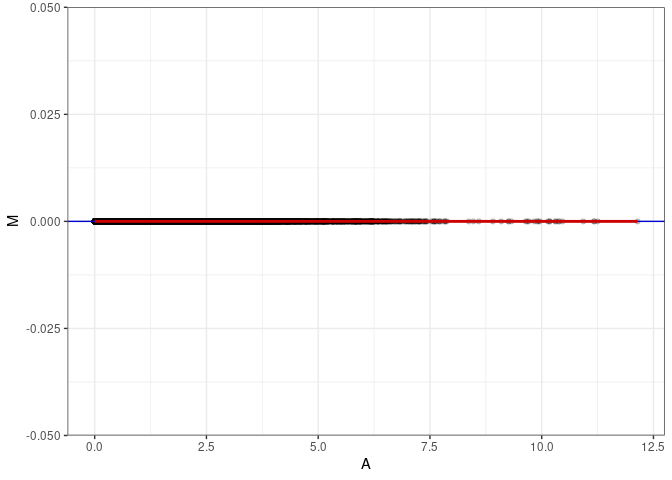
MA plot A077 - A113
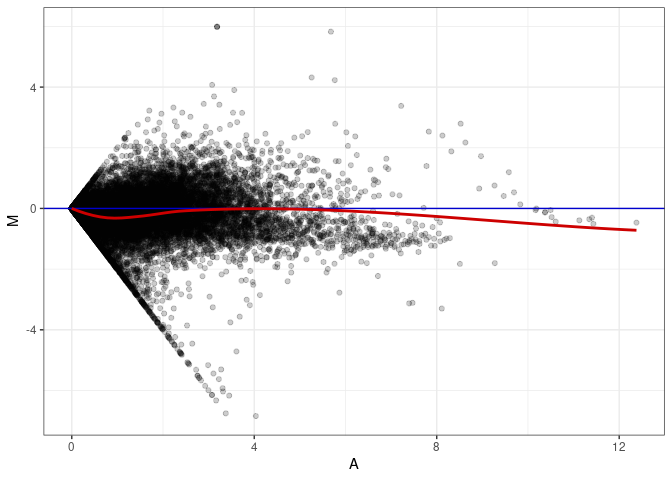
MA plot A113 - A059
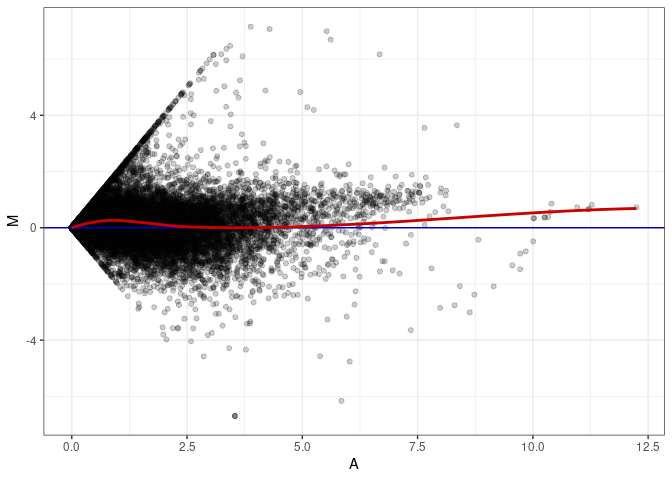
MA plot A113 - A068
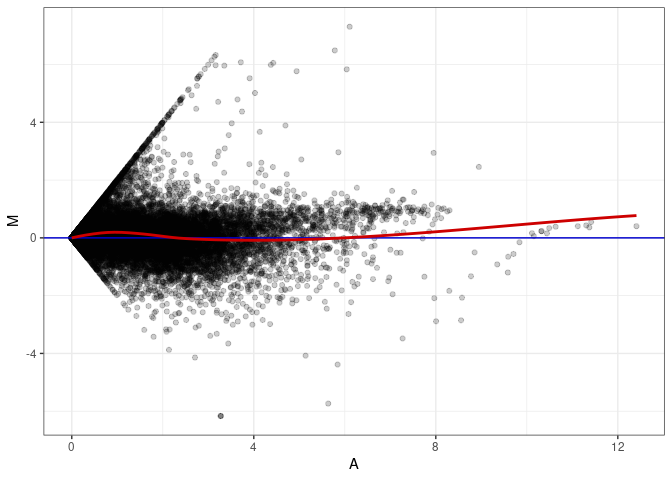
MA plot A113 - A077
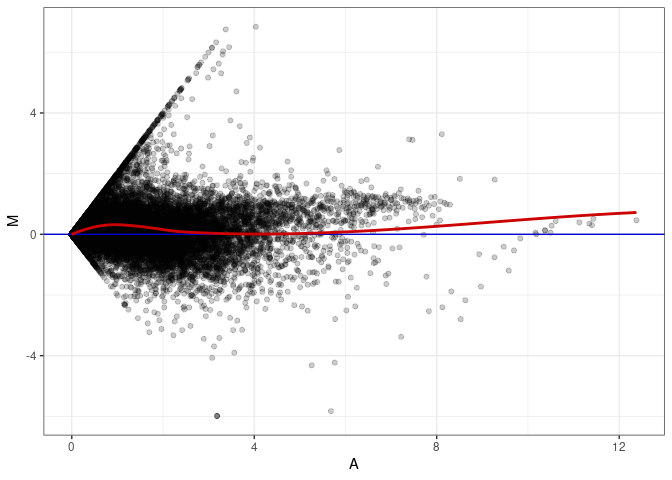
MA plot A113 - A113
MA plot for Prefrontal Cortex 2DG 4wks
subdata <- log.tdata.FPKM.sample.info %>% rownames_to_column() %>% filter(Tissue =="Pre-frontal Cortex" & Treatment == "2DG" & Time == "4 wks") %>% column_to_rownames()
subdata <- subdata[,-c(27237:27239)]
log.data.FPKM.sample.info <- t(subdata)
for(i in 1:ncol(log.data.FPKM.sample.info)){
for(j in 1:ncol(log.data.FPKM.sample.info)){
x = log.data.FPKM.sample.info[,i]
y = log.data.FPKM.sample.info[,j]
## M-values
M=x-y ## A-values
A = (x + y)/2
df = data.frame(A, M)
cat('###',"MA plot", colnames(log.data.FPKM.sample.info)[[i]], "-", colnames(log.data.FPKM.sample.info)[[j]] ,' \n')
p <- ggplot(df, aes(x = A, y = M)) + geom_point(size = 1.5, alpha = 1/5) +
geom_hline(color = "blue3", yintercept = 0) + stat_smooth(se = FALSE, method = "loess", color = "red3") + theme_bw()
print(p)
#print(htmltools::tagList(ggplotly(p)))
cat("\n \n")
}
}MA plot A027 - A027
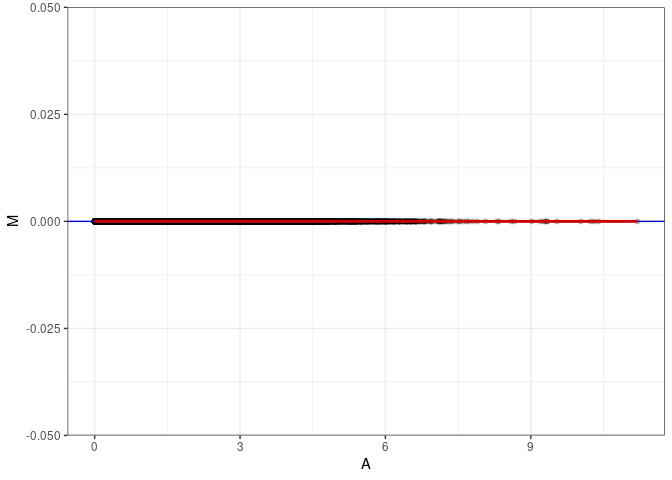
MA plot A027 - A036
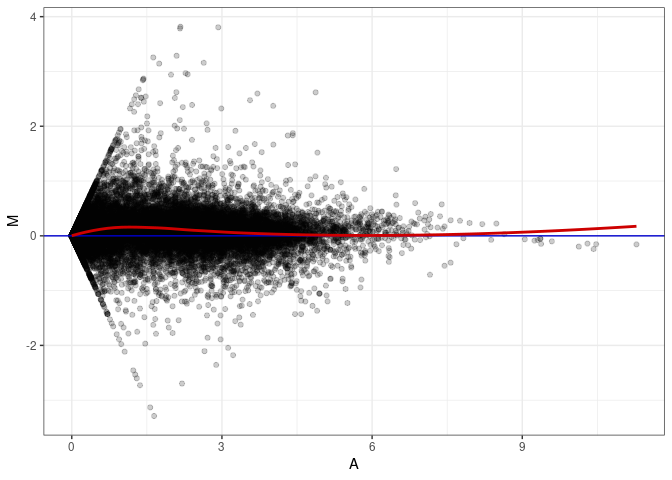
MA plot A027 - A099
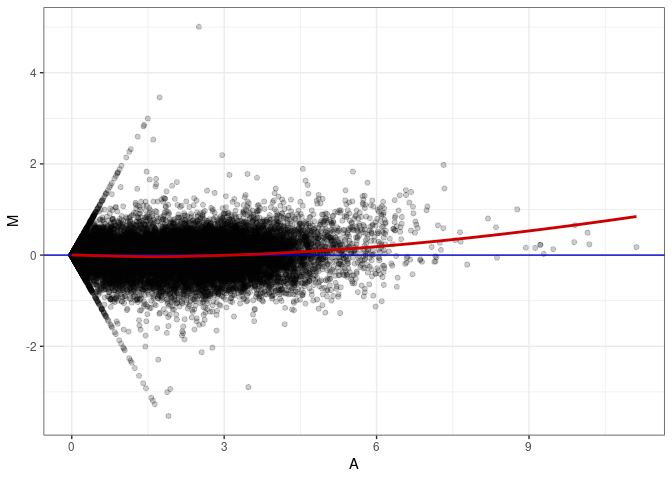
MA plot A027 - A180
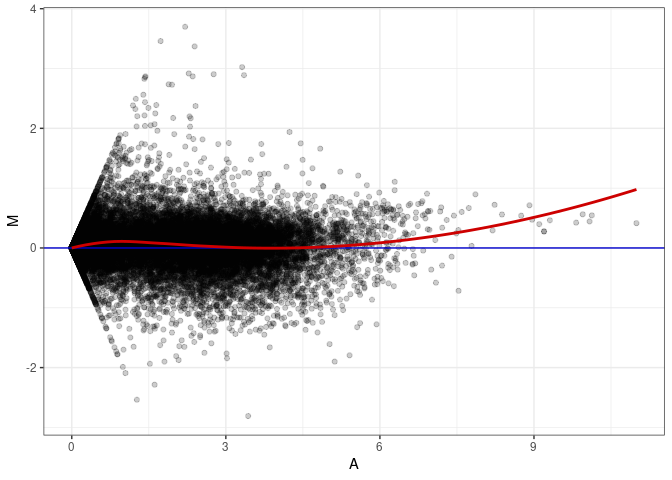
MA plot A036 - A027
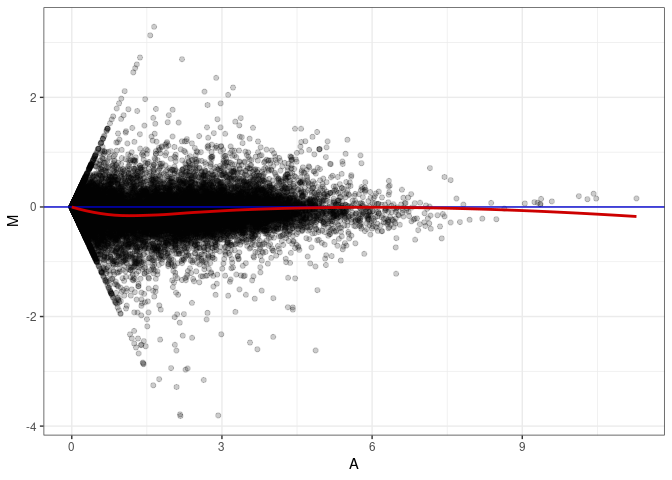
MA plot A036 - A036
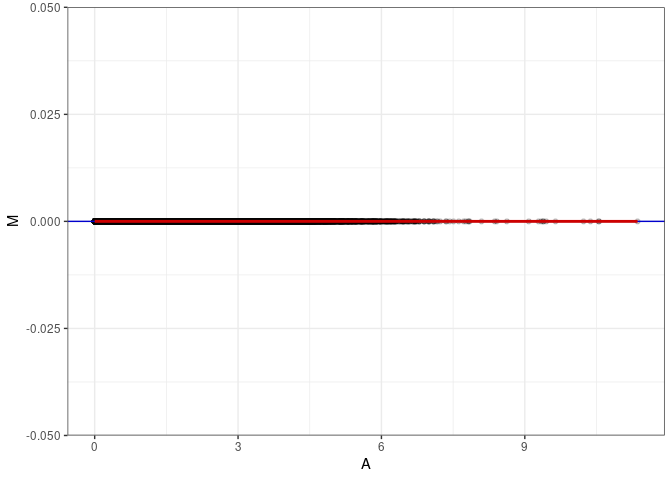
MA plot A036 - A099
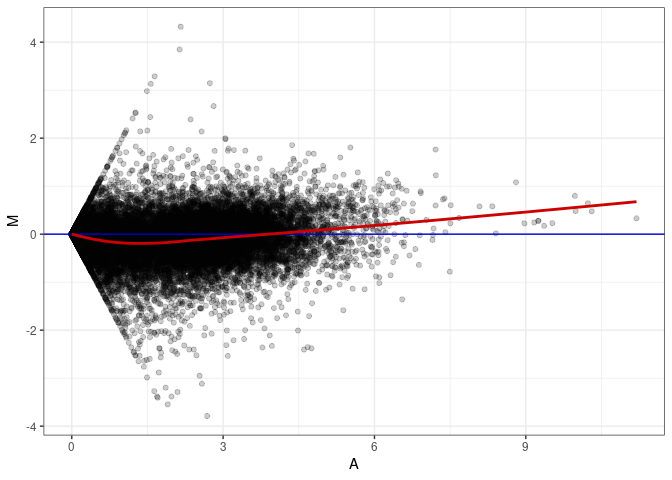
MA plot A036 - A180
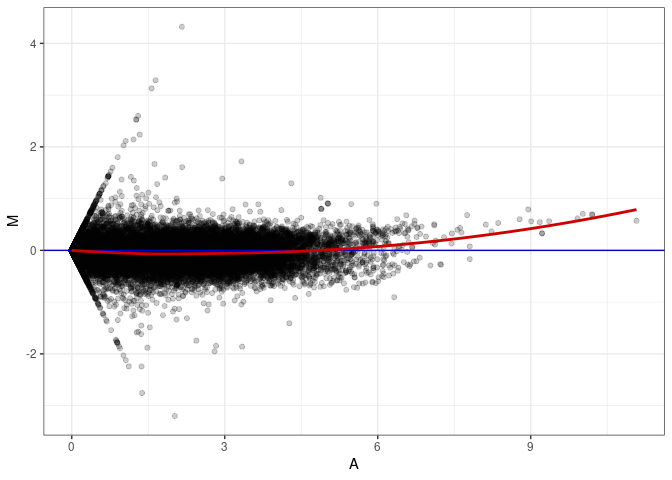
MA plot A099 - A027
MA plot A099 - A036
MA plot A099 - A099
MA plot A099 - A180
MA plot A180 - A027
MA plot A180 - A036
MA plot A180 - A099
MA plot A180 - A180
MA plot for Prefrontal Cortex control 4wks
subdata <- log.tdata.FPKM.sample.info %>% rownames_to_column() %>% filter(Tissue =="Pre-frontal Cortex" & Treatment == "None" & Time == "4 wks") %>% column_to_rownames()
subdata <- subdata[,-c(27237:27239)]
log.data.FPKM.sample.info <- t(subdata)
for(i in 1:ncol(log.data.FPKM.sample.info)){
for(j in 1:ncol(log.data.FPKM.sample.info)){
x = log.data.FPKM.sample.info[,i]
y = log.data.FPKM.sample.info[,j]
## M-values
M=x-y ## A-values
A = (x + y)/2
df = data.frame(A, M)
cat('###',"MA plot", colnames(log.data.FPKM.sample.info)[[i]], "-", colnames(log.data.FPKM.sample.info)[[j]] ,' \n')
p <- ggplot(df, aes(x = A, y = M)) + geom_point(size = 1.5, alpha = 1/5) +
geom_hline(color = "blue3", yintercept = 0) + stat_smooth(se = FALSE, method = "loess", color = "red3") + theme_bw()
print(p)
#print(htmltools::tagList(ggplotly(p)))
cat("\n \n")
}
}MA plot A009 - A009
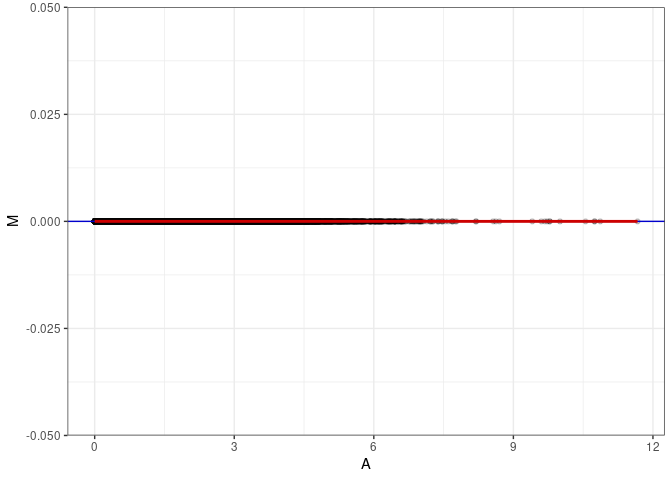
MA plot A009 - A045
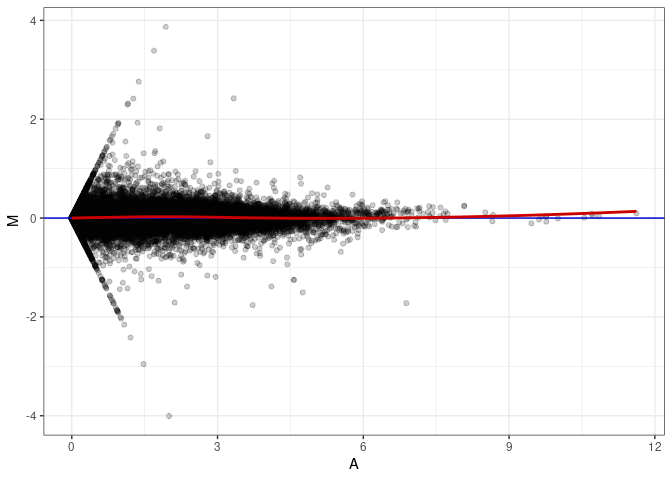
MA plot A009 - A054
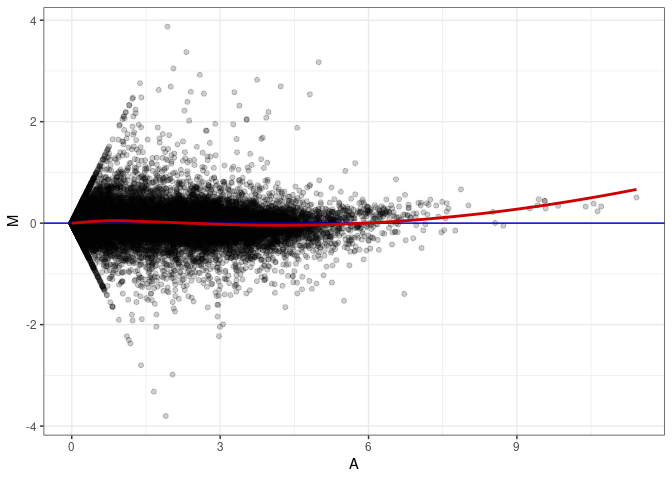
MA plot A009 - A090
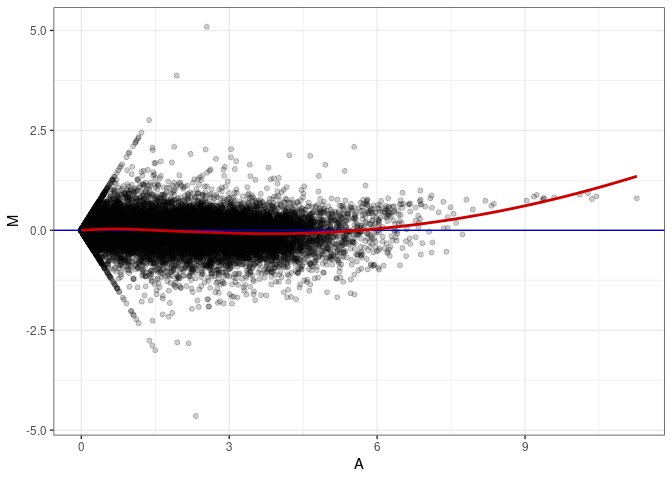
MA plot A045 - A009
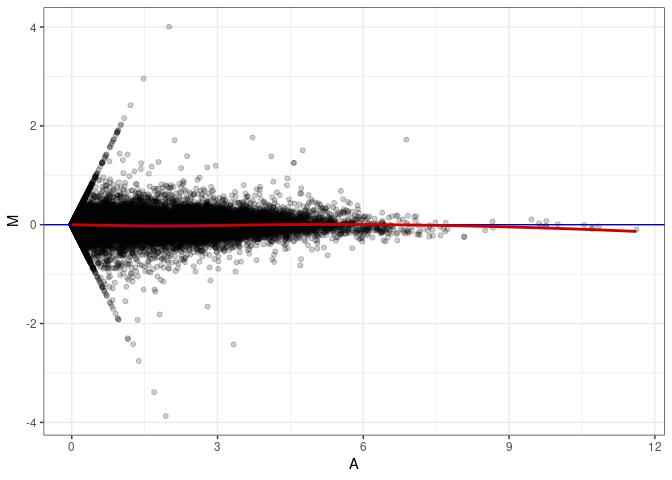
MA plot A045 - A045
MA plot A045 - A054
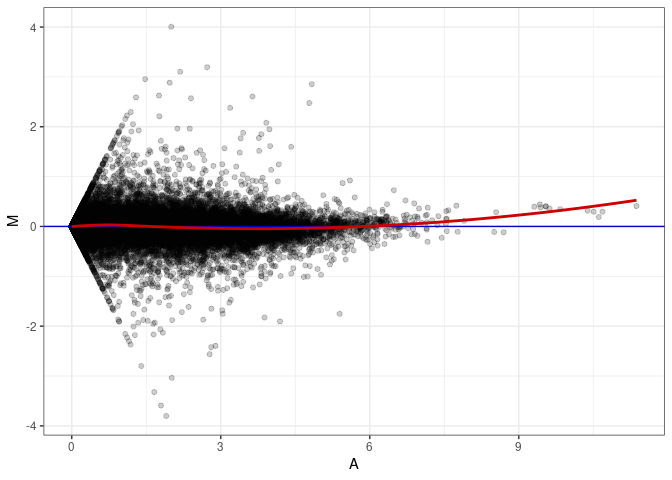
MA plot A045 - A090
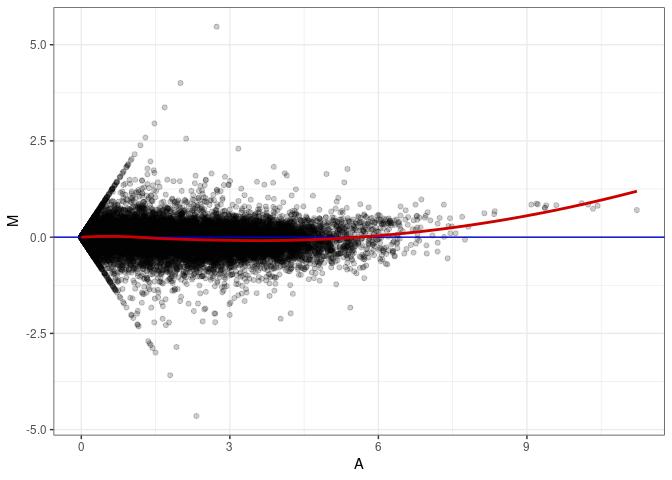
MA plot A054 - A009
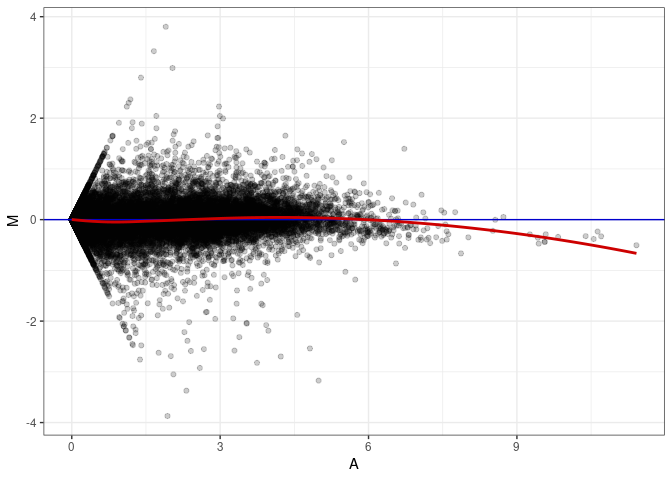
MA plot A054 - A045
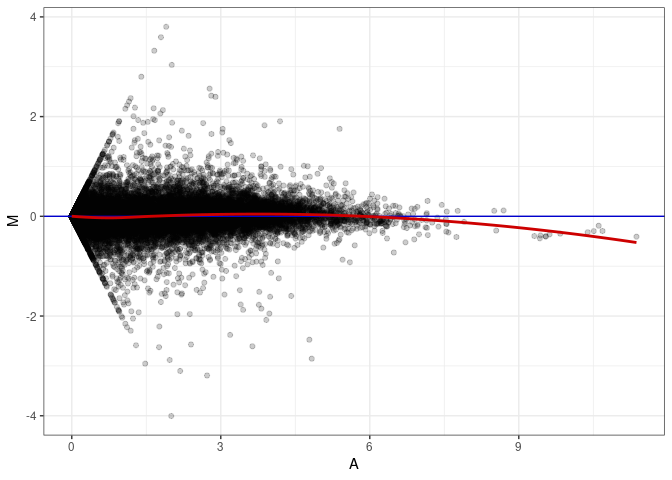
MA plot A054 - A054
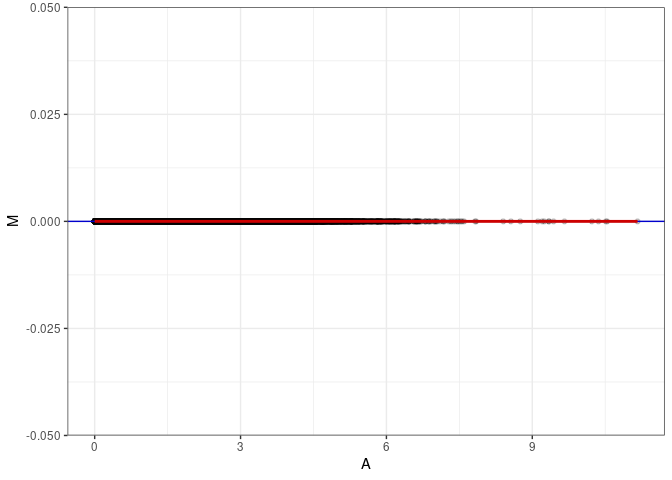
MA plot A054 - A090
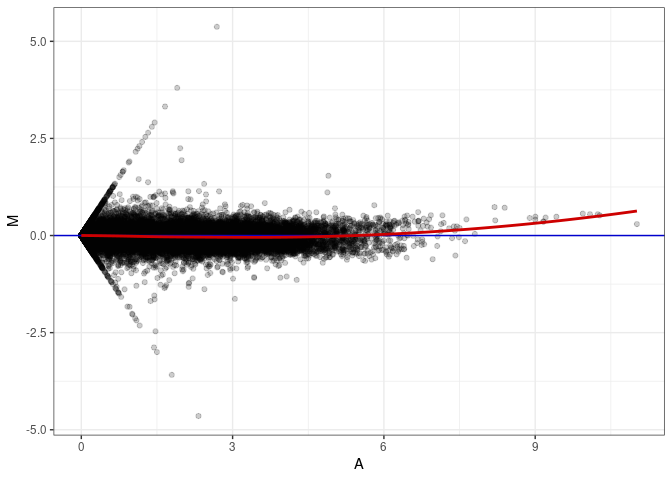
MA plot A090 - A009
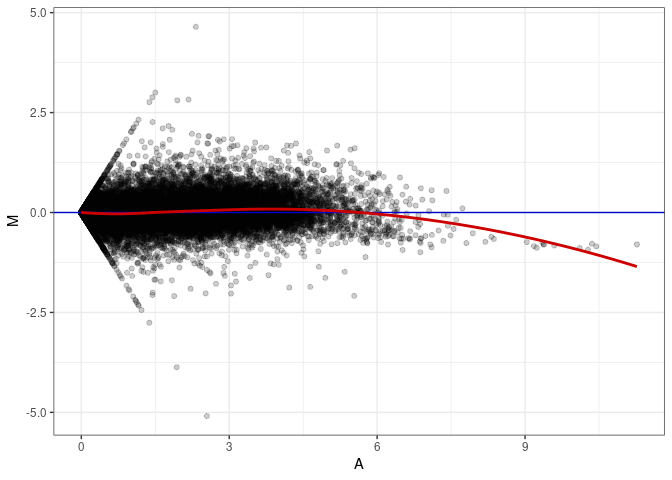
MA plot A090 - A045
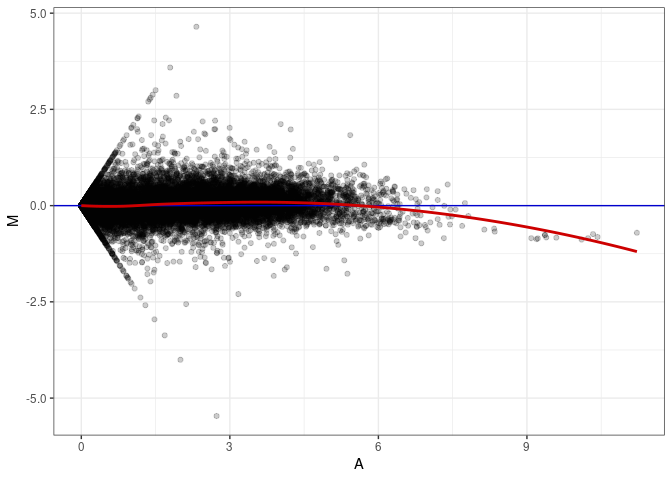
MA plot A090 - A054
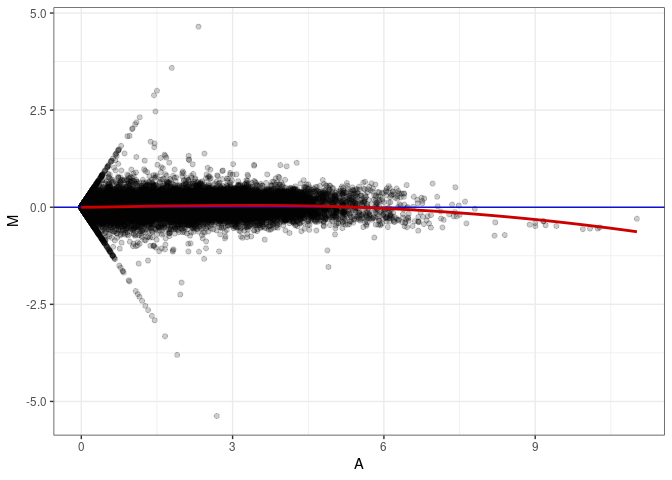
MA plot A090 - A090
MA plot for Prefrontal Cortex 2DG 96hrs
subdata <- log.tdata.FPKM.sample.info %>% rownames_to_column() %>% filter(Tissue =="Pre-frontal Cortex" & Treatment == "2DG" & Time == "96 hrs") %>% column_to_rownames()
subdata <- subdata[,-c(27237:27239)]
log.data.FPKM.sample.info <- t(subdata)
for(i in 1:ncol(log.data.FPKM.sample.info)){
for(j in 1:ncol(log.data.FPKM.sample.info)){
x = log.data.FPKM.sample.info[,i]
y = log.data.FPKM.sample.info[,j]
## M-values
M=x-y ## A-values
A = (x + y)/2
df = data.frame(A, M)
cat('###',"MA plot", colnames(log.data.FPKM.sample.info)[[i]], "-", colnames(log.data.FPKM.sample.info)[[j]] ,' \n')
p <- ggplot(df, aes(x = A, y = M)) + geom_point(size = 1.5, alpha = 1/5) +
geom_hline(color = "blue3", yintercept = 0) + stat_smooth(se = FALSE, method = "loess", color = "red3") + theme_bw()
print(p)
#print(htmltools::tagList(ggplotly(p)))
cat("\n \n")
}
}MA plot A018 - A018
MA plot A018 - A144
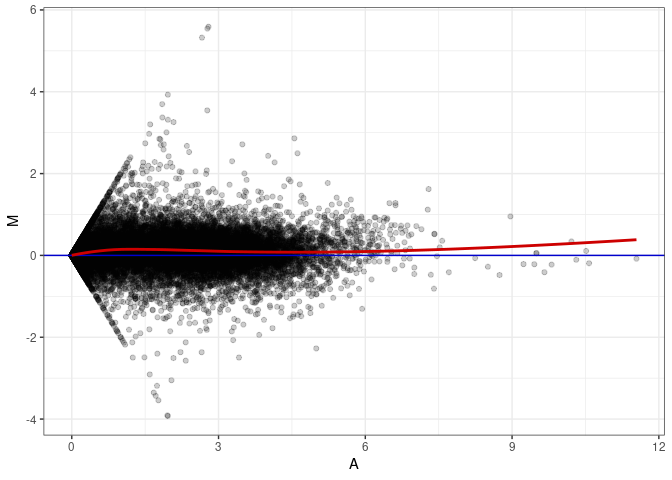
MA plot A018 - A153
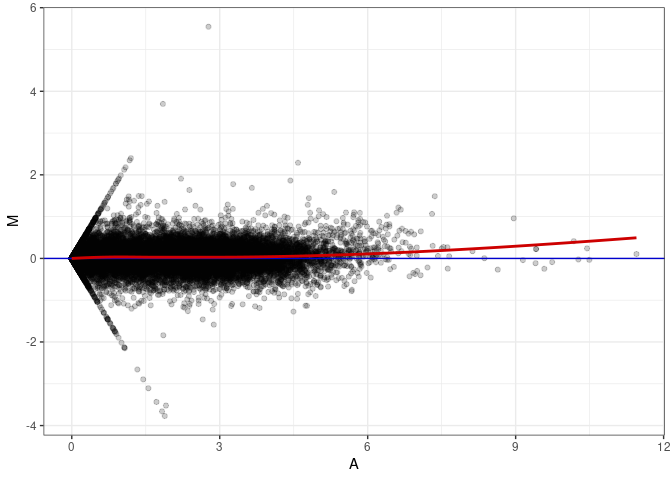
MA plot A018 - A171
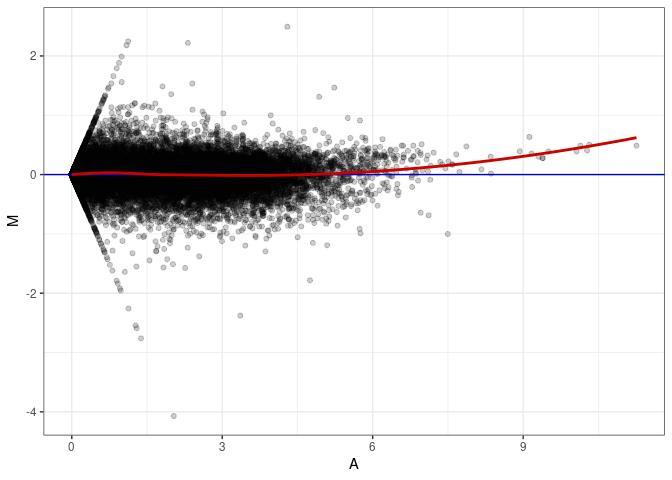
MA plot A144 - A018
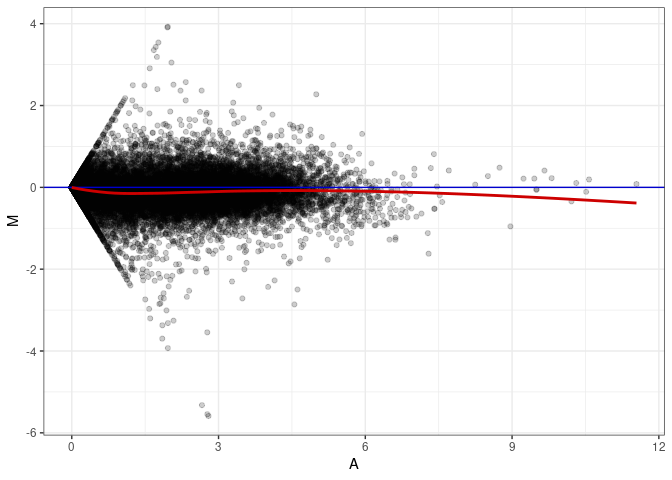
MA plot A144 - A144
MA plot A144 - A153
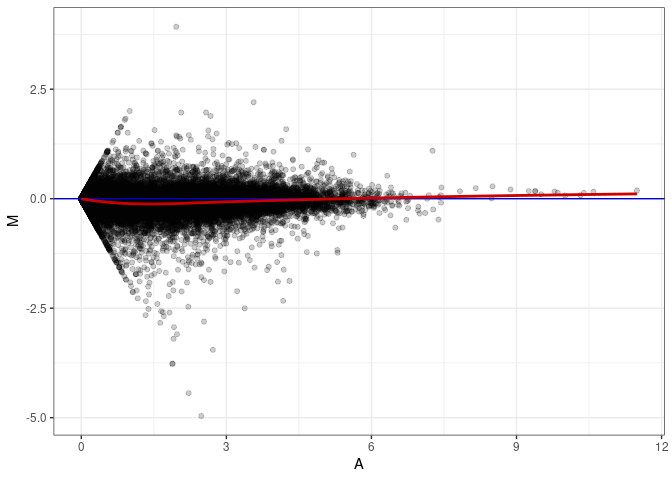
MA plot A144 - A171
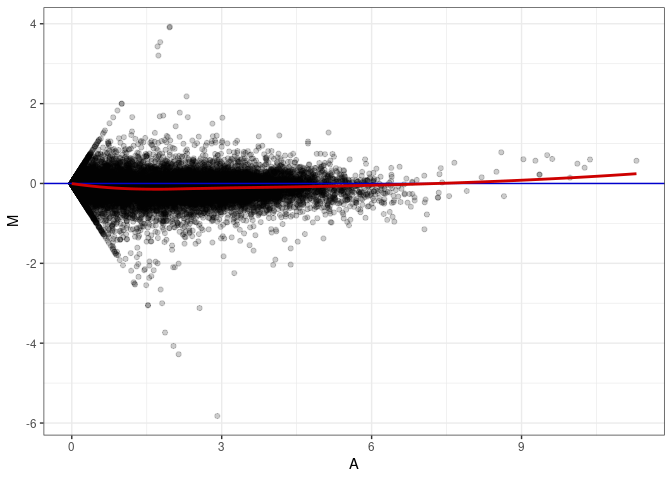
MA plot A153 - A018
MA plot A153 - A144
MA plot A153 - A153
MA plot A153 - A171
MA plot A171 - A018
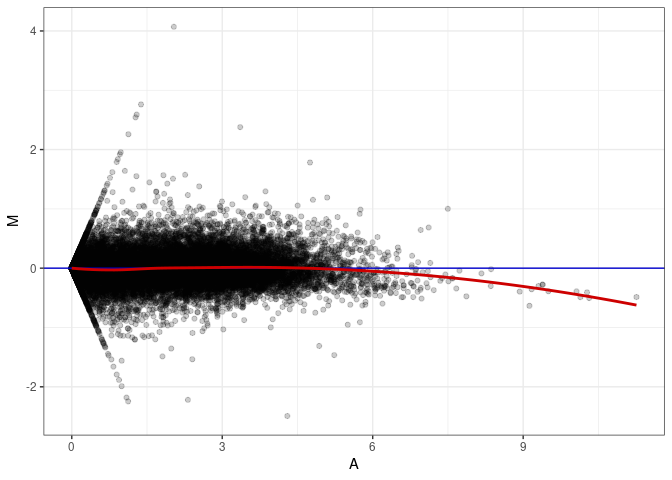
MA plot A171 - A144
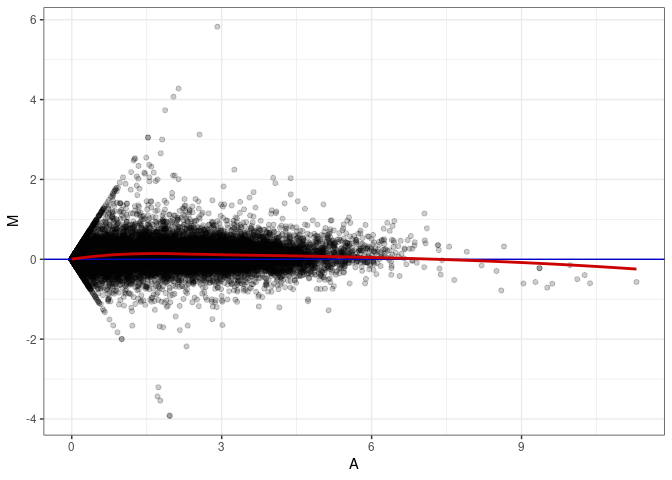
MA plot A171 - A153
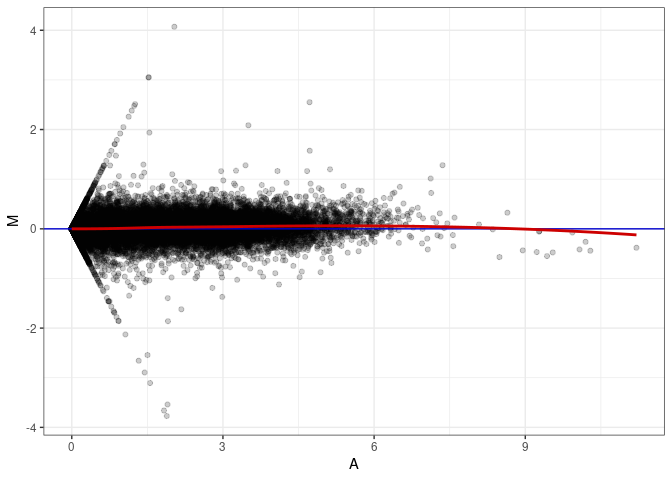
MA plot A171 - A171
MA plot for Prefrontal Cortex control 96hrs
subdata <- log.tdata.FPKM.sample.info %>% rownames_to_column() %>% filter(Tissue =="Pre-frontal Cortex" & Treatment == "None" & Time == "96 hrs") %>% column_to_rownames()
subdata <- subdata[,-c(27237:27239)]
log.data.FPKM.sample.info <- t(subdata)
for(i in 1:ncol(log.data.FPKM.sample.info)){
for(j in 1:ncol(log.data.FPKM.sample.info)){
x = log.data.FPKM.sample.info[,i]
y = log.data.FPKM.sample.info[,j]
## M-values
M=x-y ## A-values
A = (x + y)/2
df = data.frame(A, M)
cat('###',"MA plot", colnames(log.data.FPKM.sample.info)[[i]], "-", colnames(log.data.FPKM.sample.info)[[j]] ,' \n')
p <- ggplot(df, aes(x = A, y = M)) + geom_point(size = 1.5, alpha = 1/5) +
geom_hline(color = "blue3", yintercept = 0) + stat_smooth(se = FALSE, method = "loess", color = "red3") + theme_bw()
print(p)
#print(htmltools::tagList(ggplotly(p)))
cat("\n \n")
}
}MA plot A063 - A063
MA plot A063 - A072
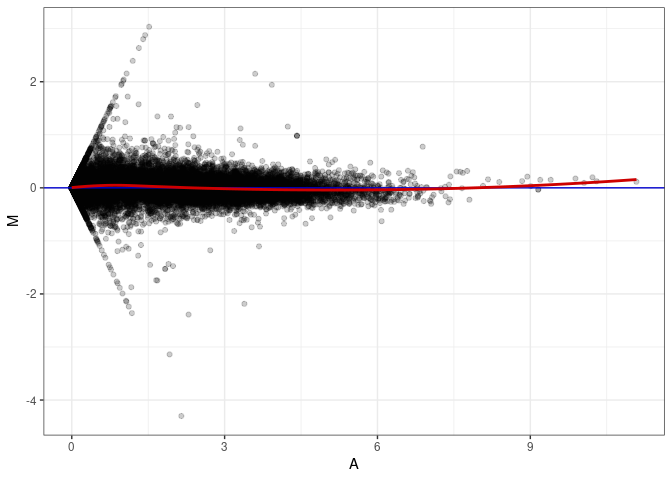
MA plot A063 - A081
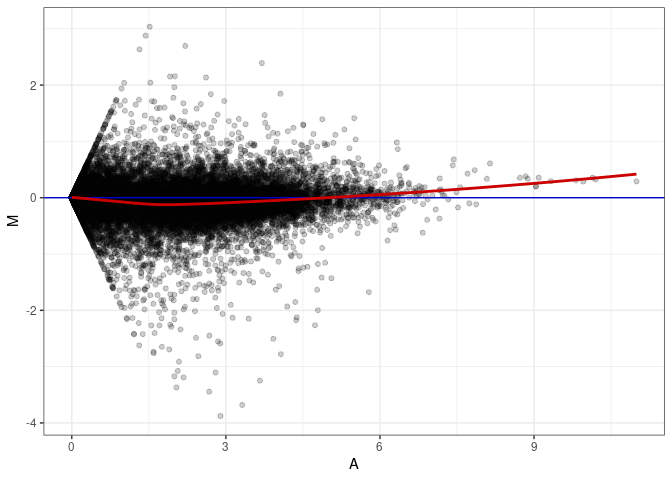
MA plot A063 - A117
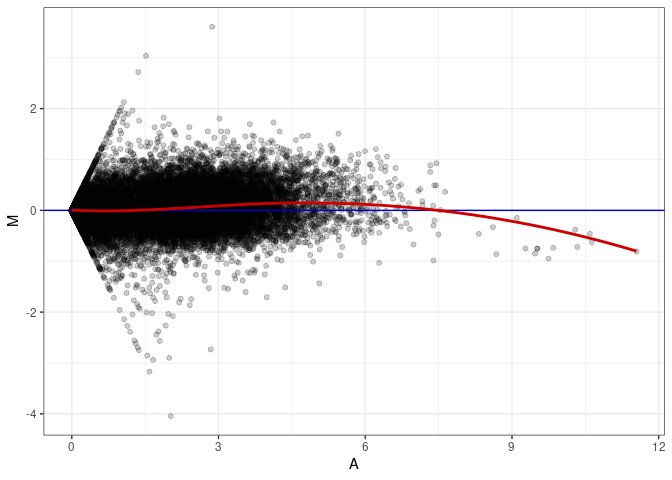
MA plot A072 - A063
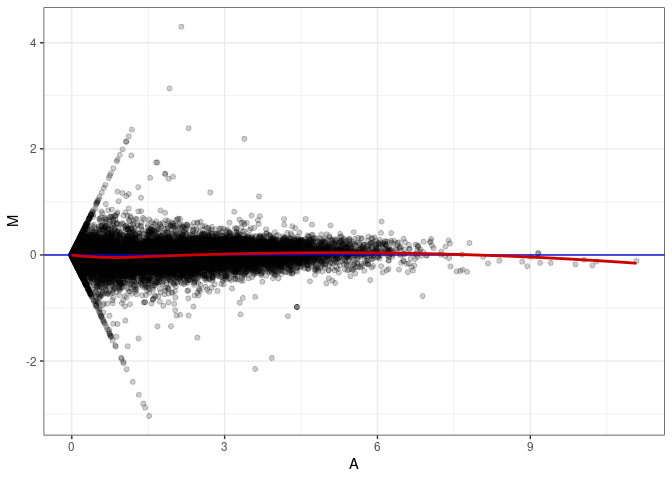
MA plot A072 - A072
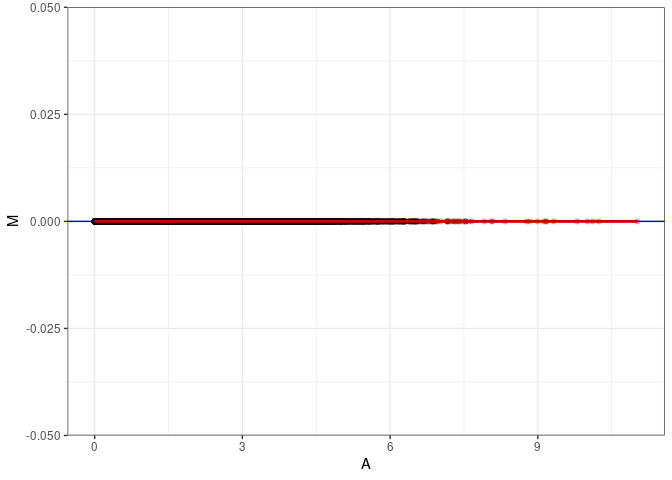
MA plot A072 - A081
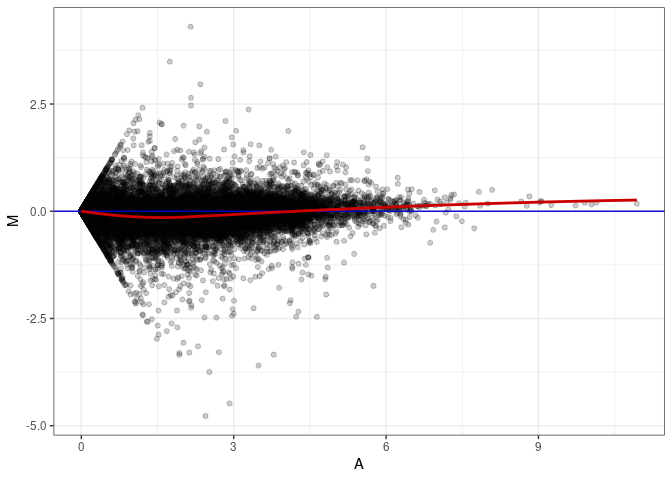
MA plot A072 - A117
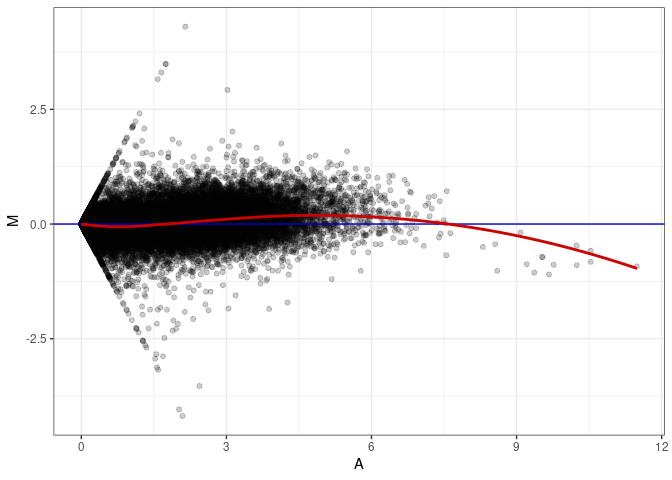
MA plot A081 - A063
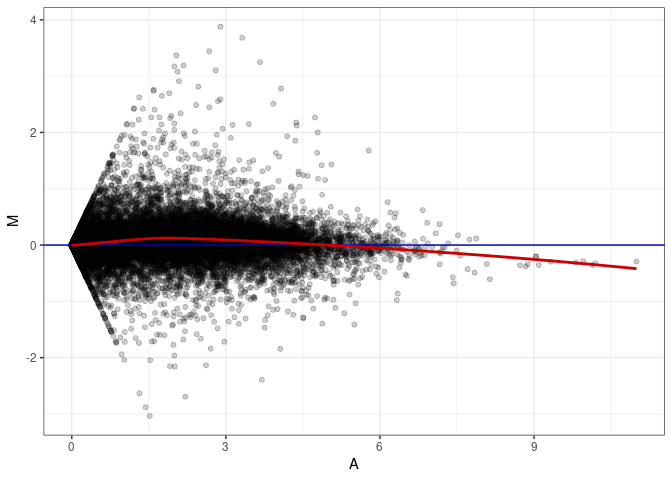
MA plot A081 - A072
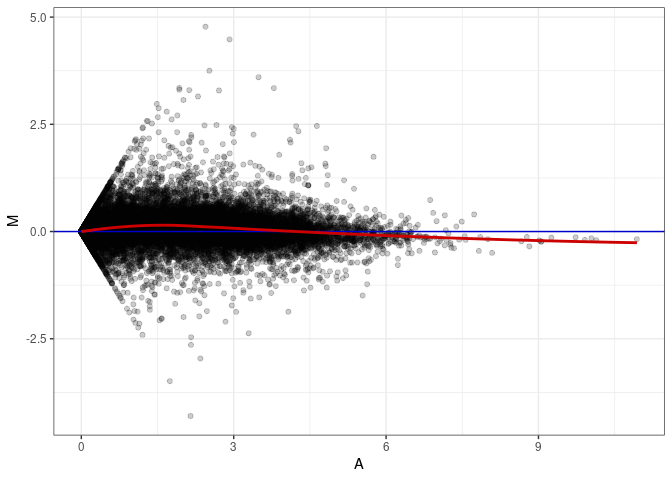
MA plot A081 - A081
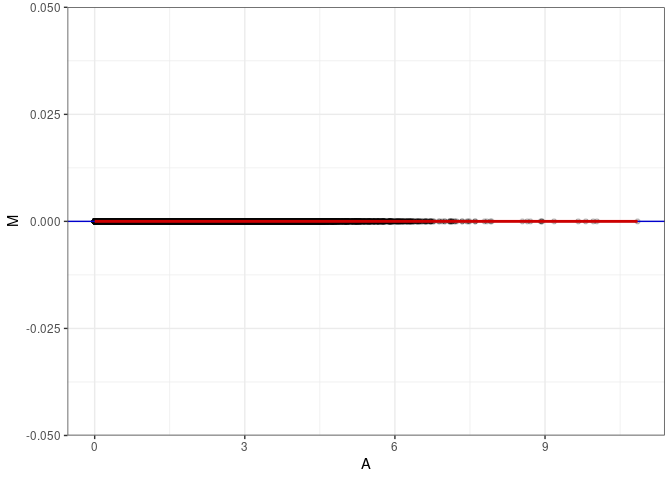
MA plot A081 - A117
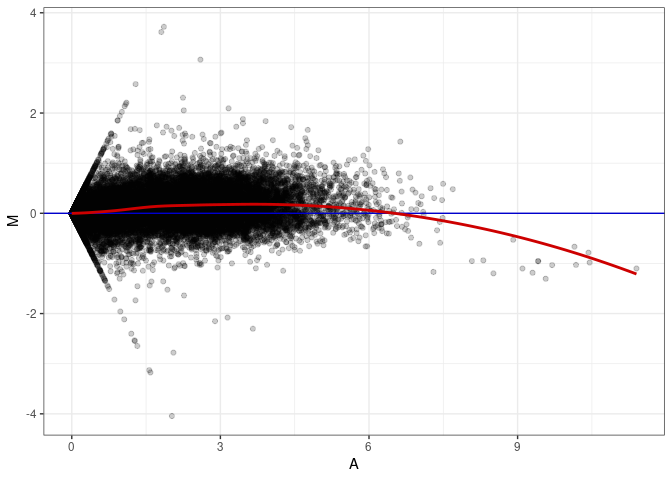
MA plot A117 - A063
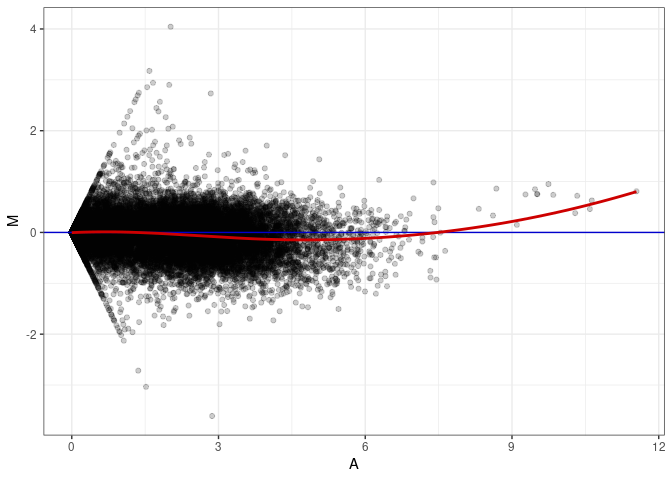
MA plot A117 - A072
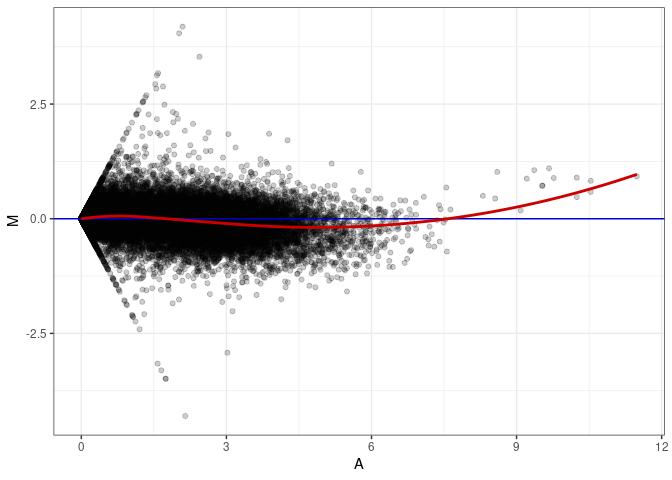
MA plot A117 - A081
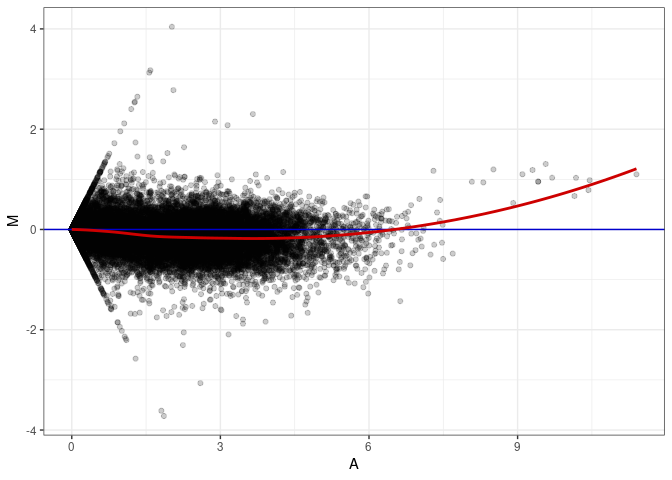
MA plot A117 - A117
Analysis performed by Ann Wells
The Carter Lab The Jackson Laboratory 2023
ann.wells@jax.org