Eigenmetabolite Stratification Prefrontal Cortex
Ann Wells
April 23, 2023
Introduction and Data files
This dataset contains nine tissues (heart, hippocampus, hypothalamus, kidney, liver, prefrontal cortex, skeletal muscle, small intestine, and spleen) from C57BL/6J mice that were fed 2-deoxyglucose (6g/L) through their drinking water for 96hrs or 4wks. 96hr mice were given their 2DG treatment 2 weeks after the other cohort started the 4 week treatment. The organs from the mice were harvested and processed for metabolomics and transcriptomics. The data in this document pertains to the transcriptomics data only. The counts that were used were FPKM normalized before being log transformed. It was determined that sample A113 had low RNAseq quality and through further analyses with PCA, MA plots, and clustering was an outlier and will be removed for the rest of the analyses performed. This document will determine the overall summary expression of each module across the main effects, as well as, assess the significance of each main effect and their interaction, using ANOVA, for each module and assess potential interactions visually for each module.
needed.packages <- c("tidyverse", "here", "functional", "gplots", "dplyr", "GeneOverlap", "R.utils", "reshape2","magrittr","data.table", "RColorBrewer","preprocessCore", "ARTool","emmeans", "phia", "gProfileR", "WGCNA","plotly", "pheatmap","pander", "kableExtra")
for(i in 1:length(needed.packages)){library(needed.packages[i], character.only = TRUE)}
source(here("source_files","WGCNA_source.R"))
source(here("source_files","plot_theme.R"))tdata.FPKM.sample.info <- readRDS(here("Data","20190406_RNAseq_B6_4wk_2DG_counts_phenotypes.RData"))
tdata.FPKM <- readRDS(here("Data","20190406_RNAseq_B6_4wk_2DG_counts_numeric.RData"))
log.tdata.FPKM <- log(tdata.FPKM + 1)
log.tdata.FPKM <- as.data.frame(log.tdata.FPKM)
log.tdata.FPKM.sample.info <- cbind(log.tdata.FPKM, tdata.FPKM.sample.info[,27238:27240])
log.tdata.FPKM.sample.info <- log.tdata.FPKM.sample.info %>% rownames_to_column() %>% filter(rowname != "A113") %>% column_to_rownames()
log.tdata.FPKM.subset <- log.tdata.FPKM[,colMeans(log.tdata.FPKM != 0) > 0.5]
log.tdata.FPKM.sample.info.subset <- cbind(log.tdata.FPKM.subset,tdata.FPKM.sample.info[,27238:27240])
log.tdata.FPKM.sample.info.subset <- log.tdata.FPKM.sample.info.subset %>% rownames_to_column() %>% filter(rowname != "A113") %>% column_to_rownames()
log.tdata.FPKM.sample.info.subset.prefrontal.cortex <- log.tdata.FPKM.sample.info.subset %>% rownames_to_column() %>% filter(Tissue == "Pre-frontal Cortex") %>% column_to_rownames()
log.tdata.FPKM.sample.info.subset.prefrontal.cortex$Treatment[log.tdata.FPKM.sample.info.subset.prefrontal.cortex$Treatment=="None"] <- "Control"
log.tdata.FPKM.sample.info.subset.prefrontal.cortex <- cbind(log.tdata.FPKM.sample.info.subset.prefrontal.cortex, Time.Treatment = paste(log.tdata.FPKM.sample.info.subset.prefrontal.cortex$Time, log.tdata.FPKM.sample.info.subset.prefrontal.cortex$Treatment))
module.labels <- readRDS(here("Data","Prefrontal Cortex","log.tdata.FPKM.sample.info.subset.prefrontal.cortex.WGCNA.module.labels.RData"))
module.eigens <- readRDS(here("Data","Prefrontal Cortex","log.tdata.FPKM.sample.info.subset.prefrontal.cortex.WGCNA.module.eigens.RData"))
modules <- readRDS(here("Data","Prefrontal Cortex","log.tdata.FPKM.sample.info.subset.prefrontal.cortex.WGCNA.module.membership.RData"))
net.deg <- readRDS(here("Data","Prefrontal Cortex","Chang_2DG_BL6_connectivity_prefrontal_cortex.RData"))
ensembl.location <- readRDS(here("Data","Ensembl_gene_id_and_location.RData"))Eigengene Stratification
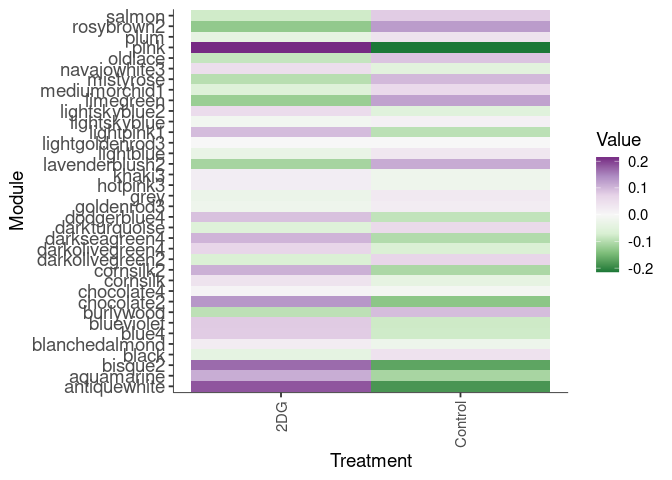
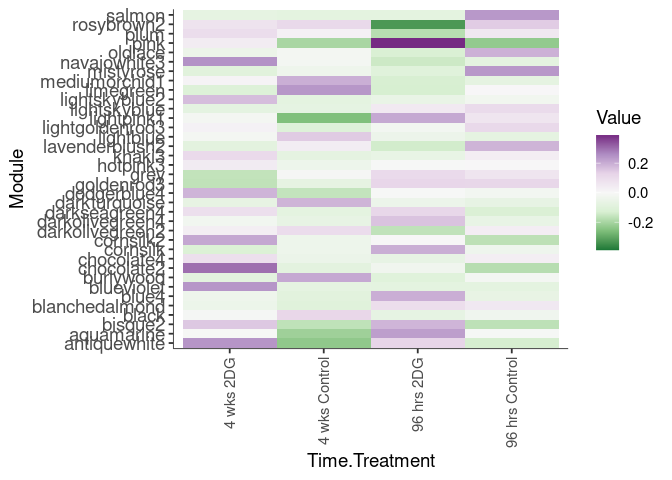
Eigengene were stratified by time and treatment. The heatmap is a matrix of the average eigengene value for each level of the trait.
factors <- c("Time","Treatment","Time.Treatment")
eigenmetabolite(factors,log.tdata.FPKM.sample.info.subset.prefrontal.cortex)Time
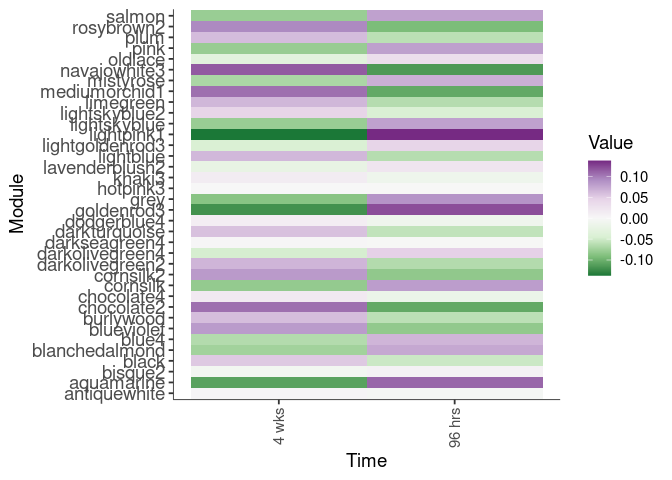
Treatment
Time.Treatment
ANOVA
An ANOVA using aligned rank transformation was performed for each module. The full model is y ~ time + treatment + time:treatment.
# Three-Way ANOVA for each Eigenmetabolite
model.data = dplyr::bind_cols(module.eigens[rownames(log.tdata.FPKM.sample.info.subset.prefrontal.cortex),], log.tdata.FPKM.sample.info.subset.prefrontal.cortex[,c("Time","Treatment")])
model.data$Time <- as.factor(model.data$Time)
model.data$Treatment <- as.factor(model.data$Treatment)
final.anova <- list()
for (m in colnames(module.eigens)) {
a <- art(data = model.data, model.data[,m] ~ Time*Treatment)
model <- anova(a)
adjust <- p.adjust(model$`Pr(>F)`, method = "BH")
final.anova[[m]] <- cbind(model, adjust)
}Blanchedalmond Module
DT.table(final.anova[[1]])Chocolate4 Module
DT.table(final.anova[[2]])Black Module
DT.table(final.anova[[3]])lightgoldenrod3 Module
DT.table(final.anova[[4]])Darkolivegreen2 Module
DT.table(final.anova[[5]])Aquamarine Module
DT.table(final.anova[[6]])Bisque2 Module
DT.table(final.anova[[7]])Darkolivegreen4 Module
DT.table(final.anova[[8]])Grey Module
DT.table(final.anova[[9]])Blueviolet Module
DT.table(final.anova[[10]])Darkturquoise Module
DT.table(final.anova[[11]])Khaki3 Module
DT.table(final.anova[[12]])Dodgerblue4 Module
DT.table(final.anova[[13]])Cornsilk2 Module
DT.table(final.anova[[14]])Darkseagreen4 Module
DT.table(final.anova[[15]])Lightpink1 Module
DT.table(final.anova[[16]])Antiquewhite Module
DT.table(final.anova[[18]])Pink Module
DT.table(final.anova[[19]])Salmon Module
DT.table(final.anova[[20]])Lavenderblush2 Module
DT.table(final.anova[[21]])Plum Module
DT.table(final.anova[[22]])Goldenrod3 Module
DT.table(final.anova[[23]])Chocolate2 Module
DT.table(final.anova[[24]])Blue4 Module
DT.table(final.anova[[25]])Mistyrose Module
DT.table(final.anova[[26]])Cornsilk Module
DT.table(final.anova[[27]])Lightskyblue2 Module
DT.table(final.anova[[28]])Burlywood Module
DT.table(final.anova[[29]])Oldlace Module
DT.table(final.anova[[30]])Lightblue Module
DT.table(final.anova[[31]])Limegreen Module
DT.table(final.anova[[32]])Hotpink3 Module
DT.table(final.anova[[33]])Lightskyblue Module
DT.table(final.anova[[34]])Mediumorchid1 Module
DT.table(final.anova[[35]])Rosybrown2 Module
DT.table(final.anova[[36]])Interaction plots
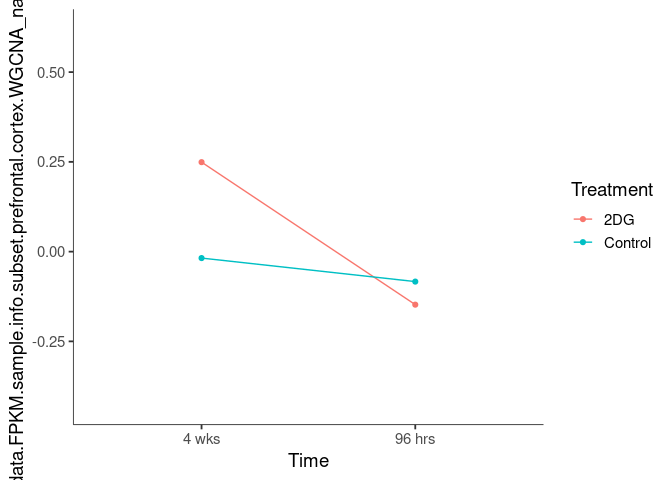
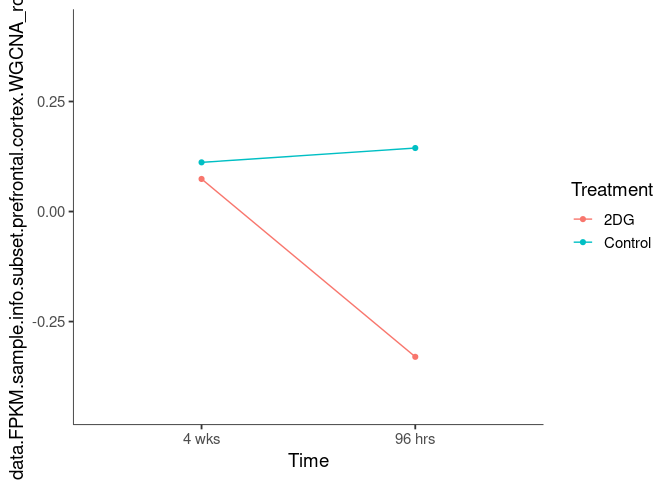
Interaction plots were created to identify which modules have a potential interaction between time and treatment. A potential interaction is identified when the two lines cross.
for (m in module.labels) {
p = plot.interaction(model.data, "Time", "Treatment", resp = m)
name <- sapply(str_split(m,"_"),"[",2)
cat("\n###",name,"\n")
print(p)
cat("\n \n")
}blanchedalmond
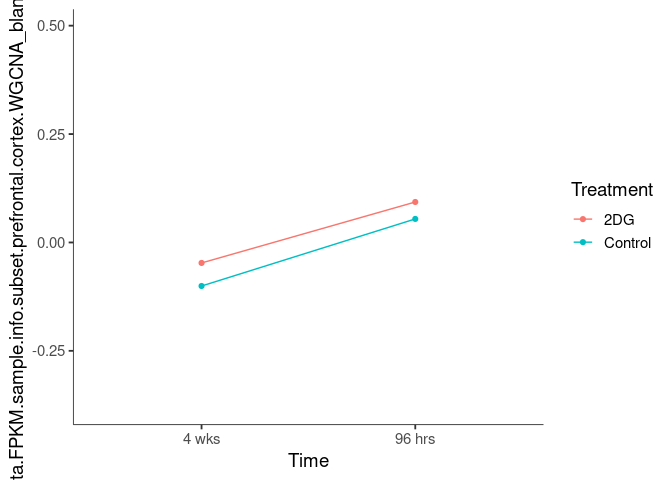
chocolate4
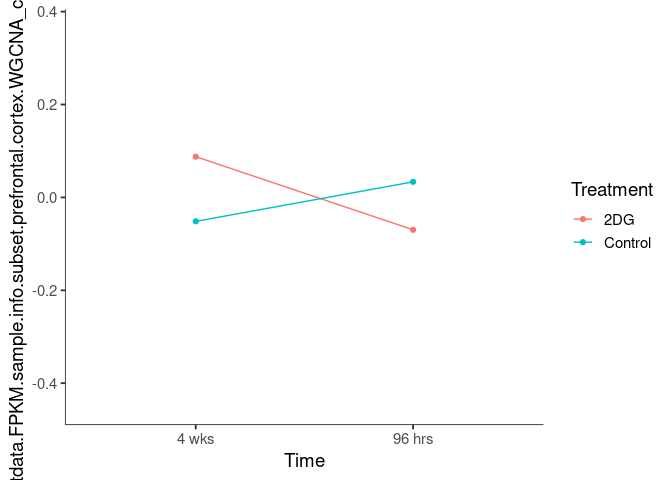
black
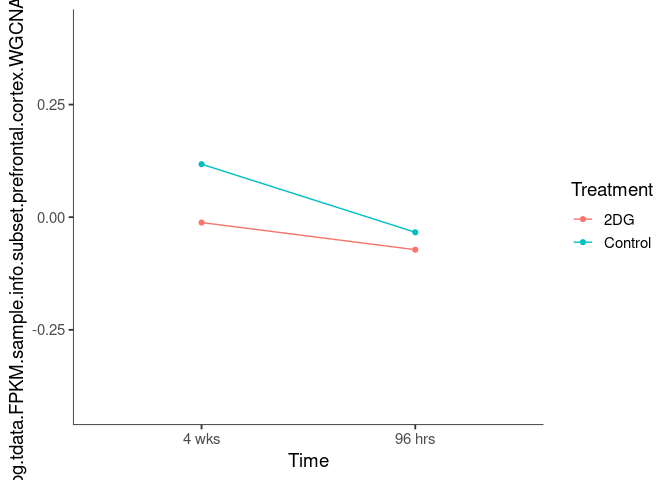
lightgoldenrod3
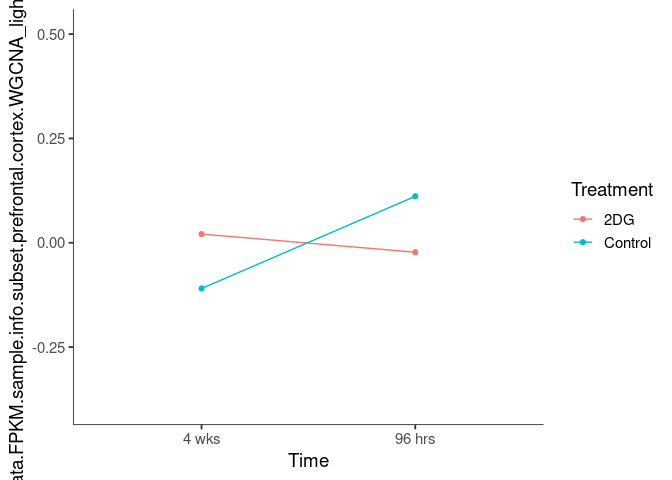
darkolivegreen2
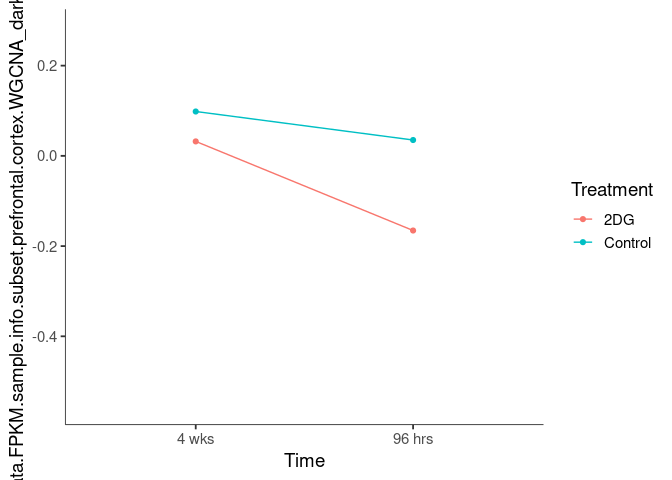
aquamarine
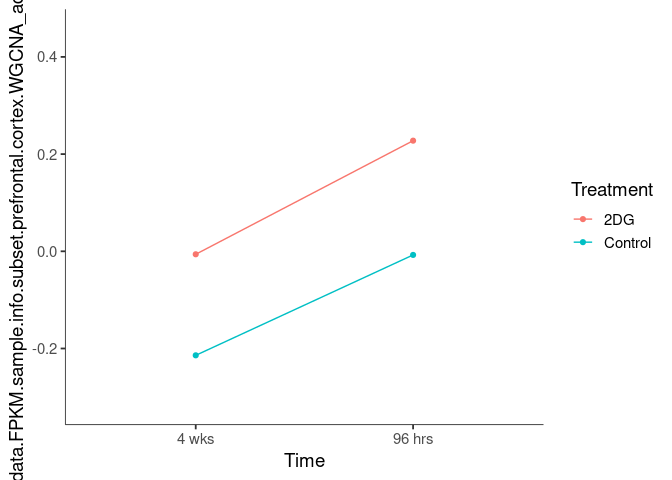
bisque2
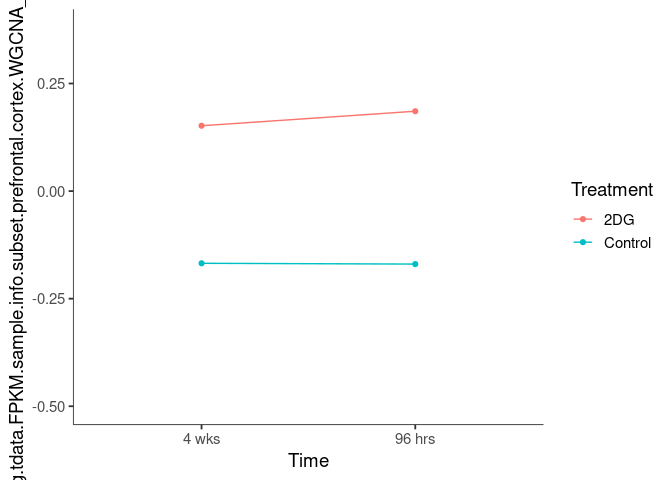
darkolivegreen4
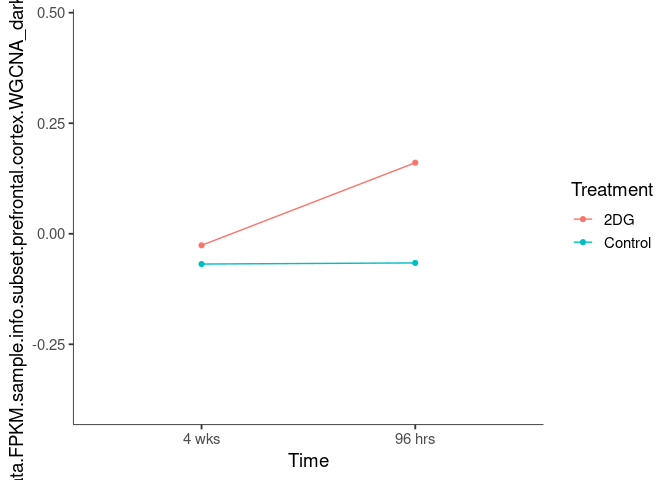
grey
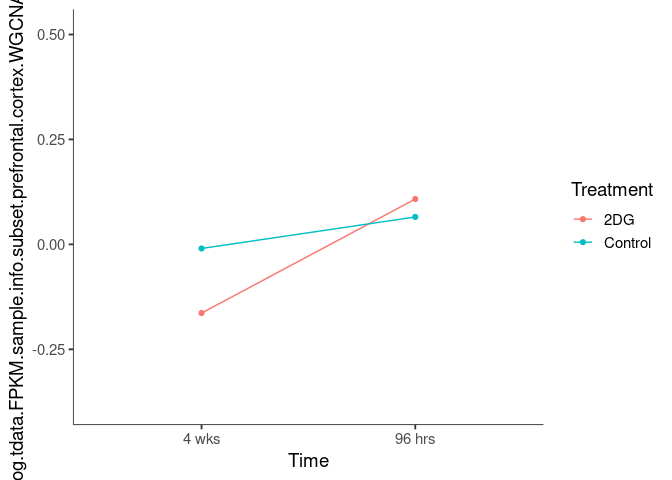
blueviolet
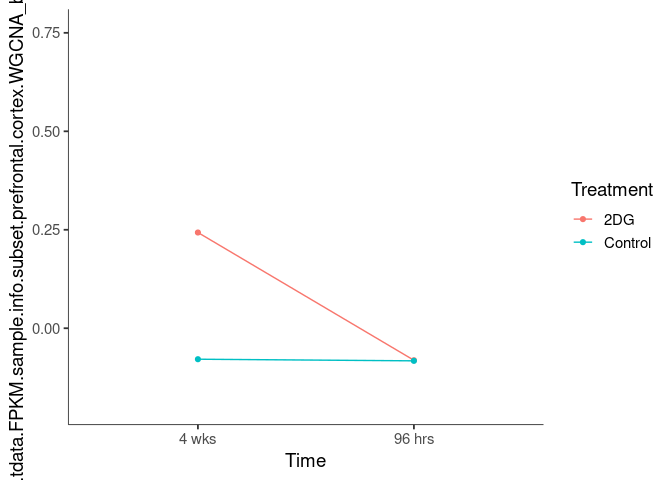
darkturquoise
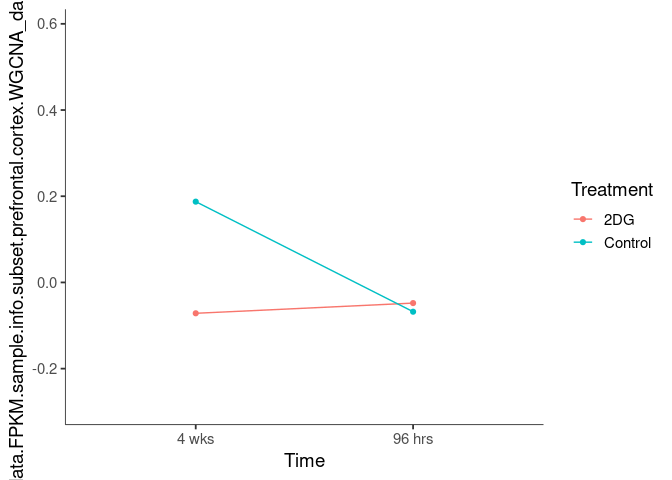
khaki3
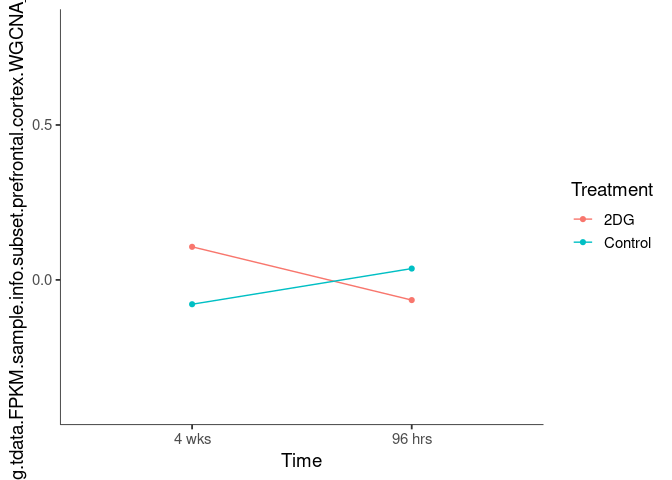
dodgerblue4
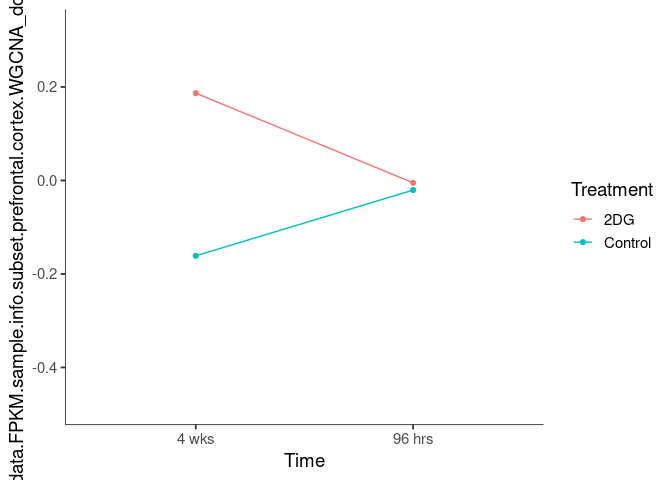
cornsilk2
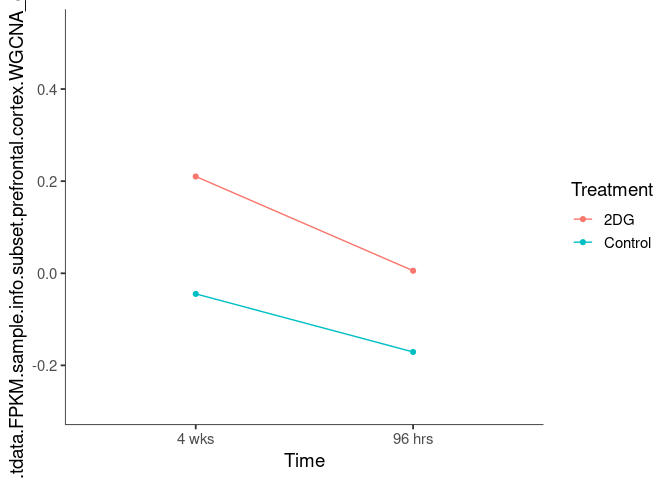
darkseagreen4
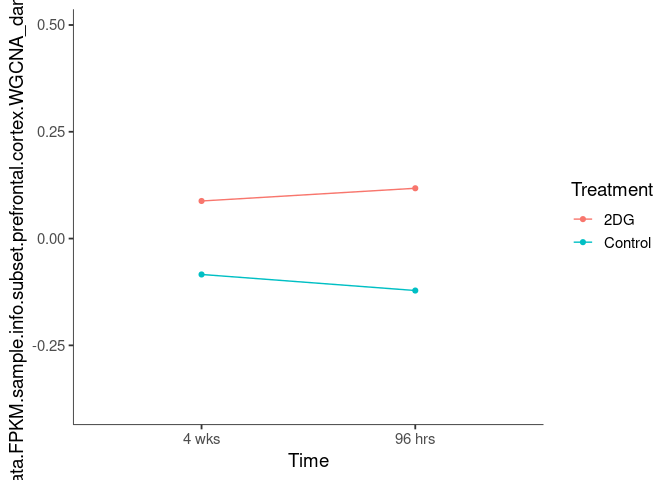
lightpink1
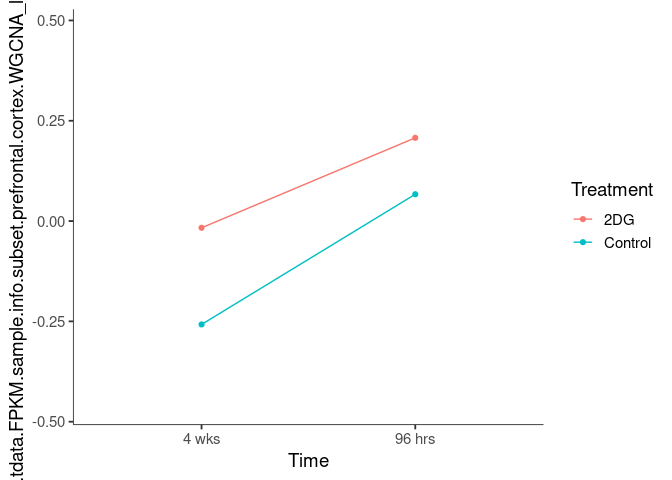
antiquewhite
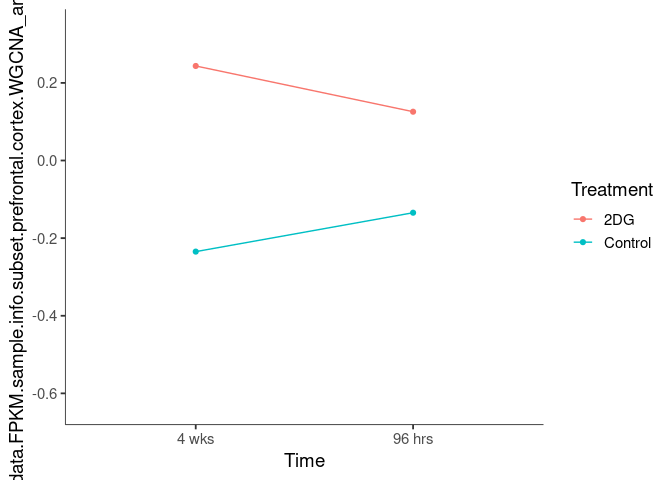
pink
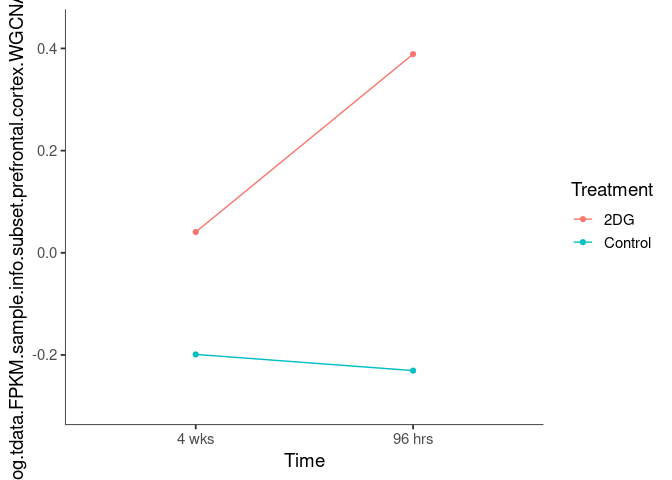
salmon
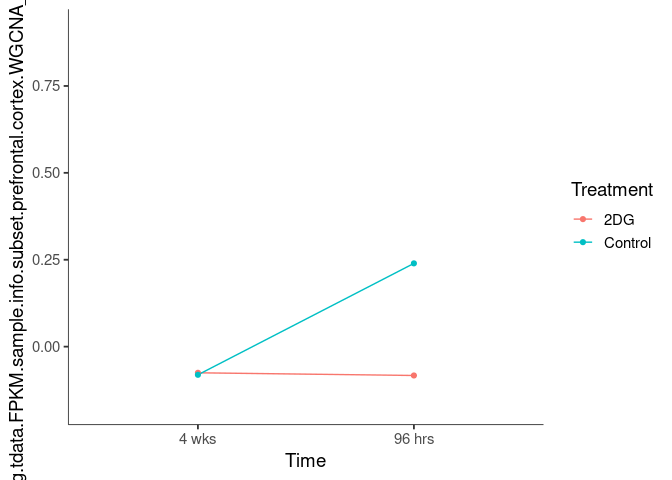
lavenderblush2
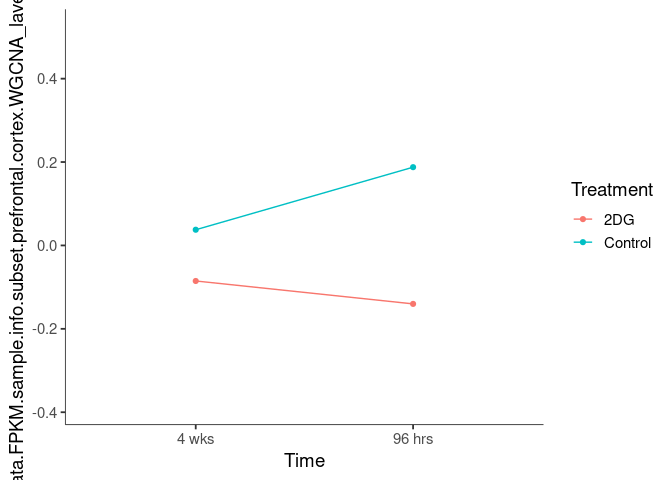
plum
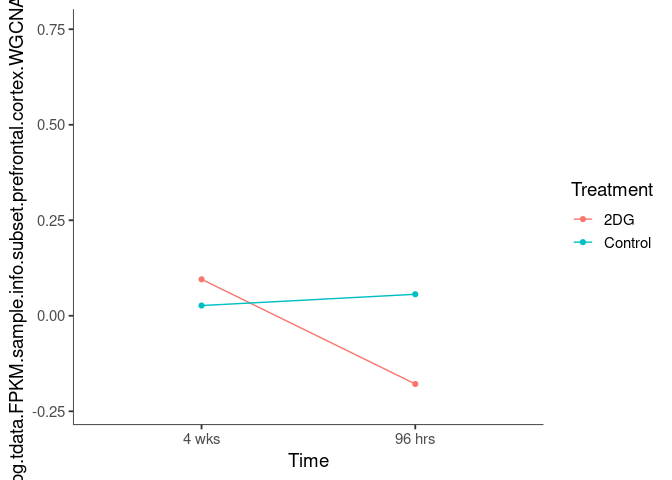
goldenrod3
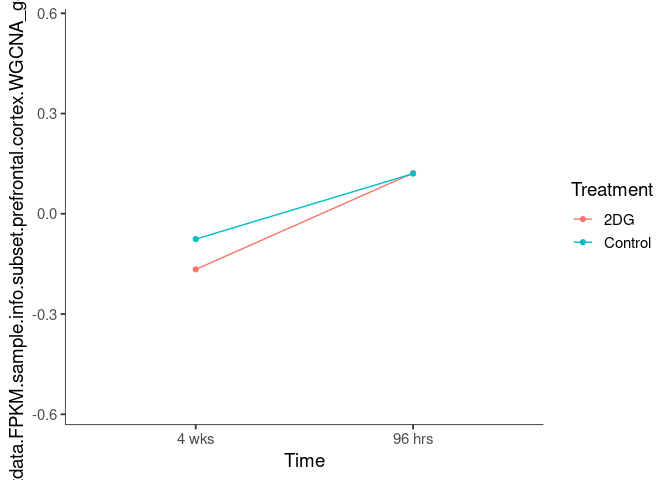
chocolate2
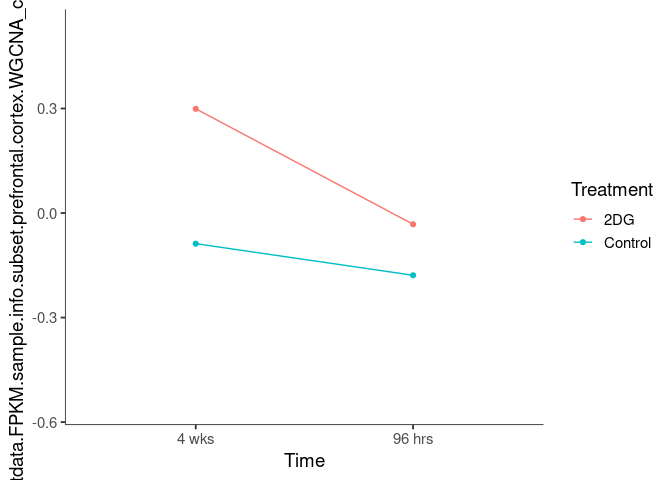
blue4
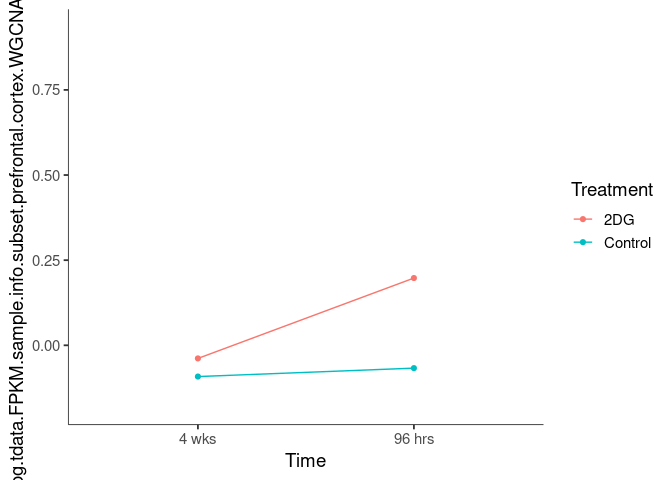
mistyrose
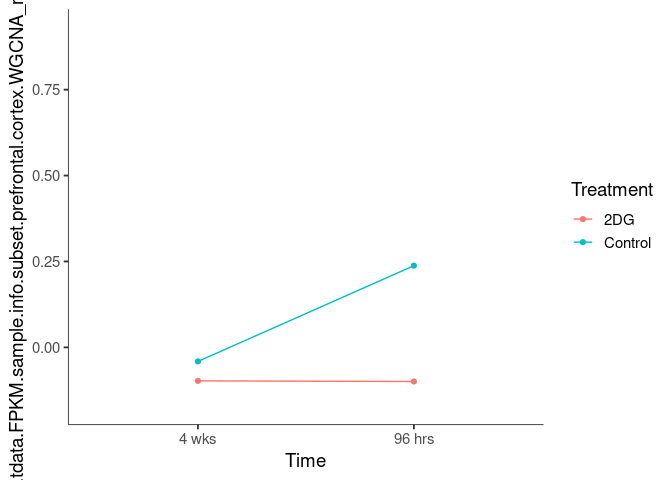
cornsilk
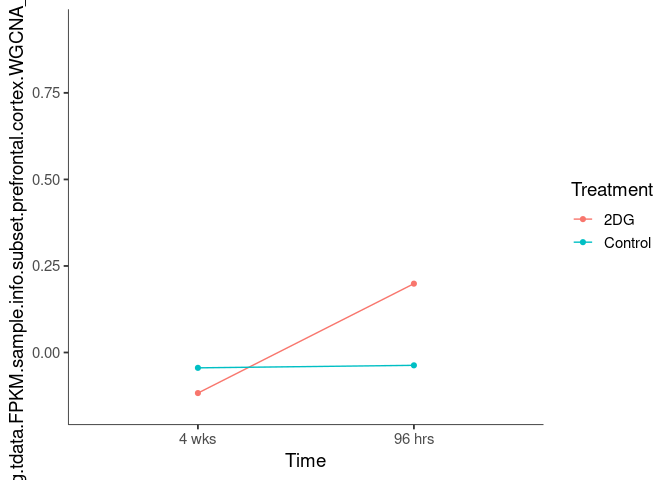
lightskyblue2
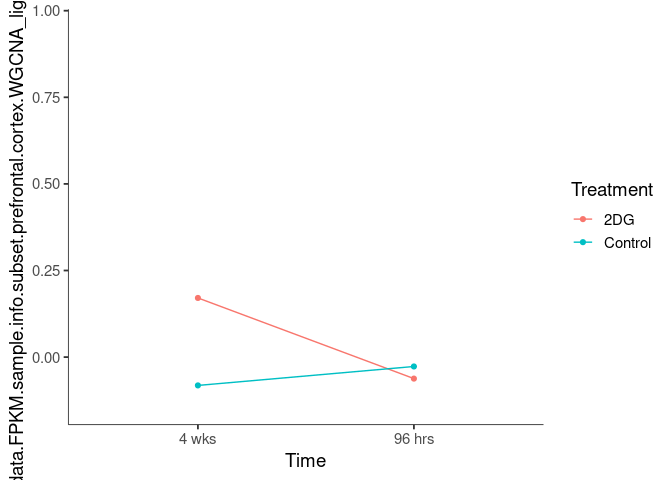
burlywood
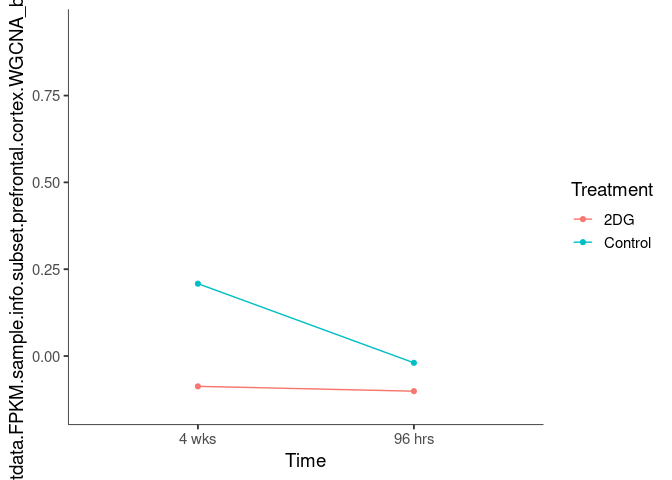
oldlace
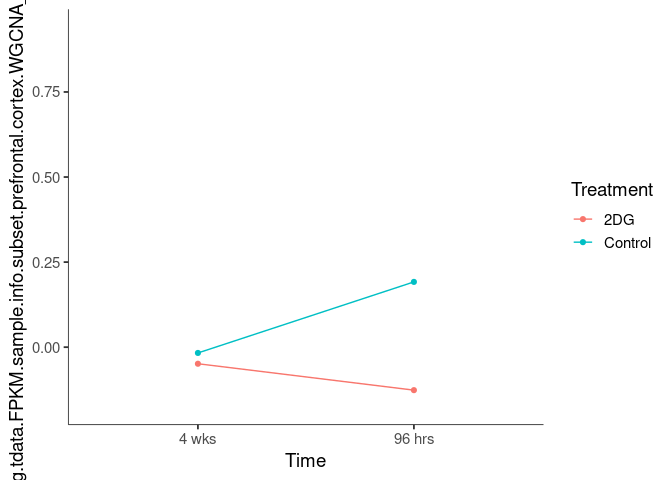
lightblue
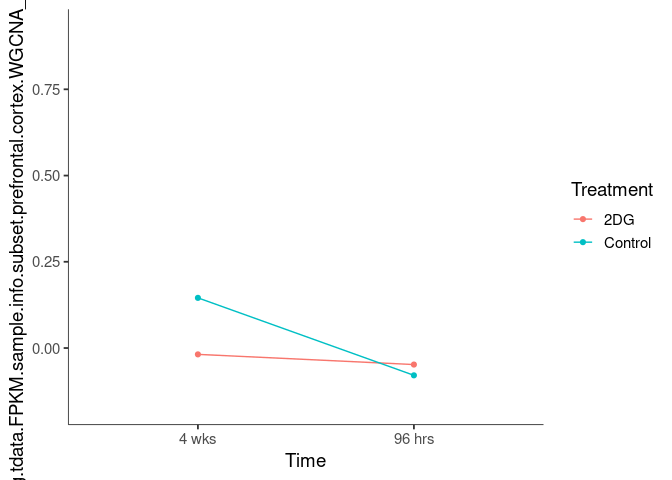
limegreen
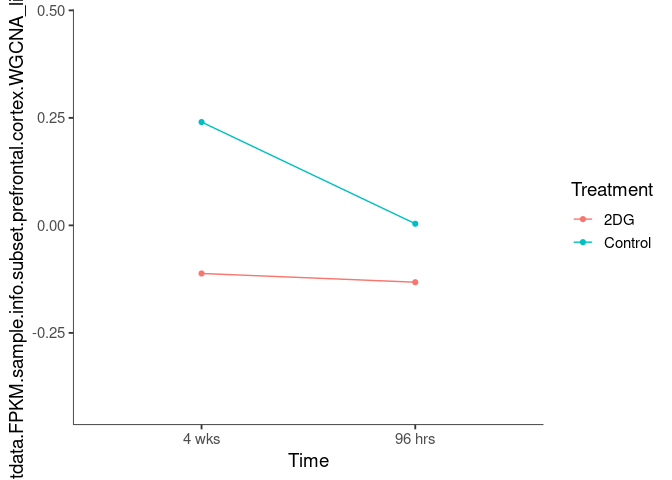
hotpink3
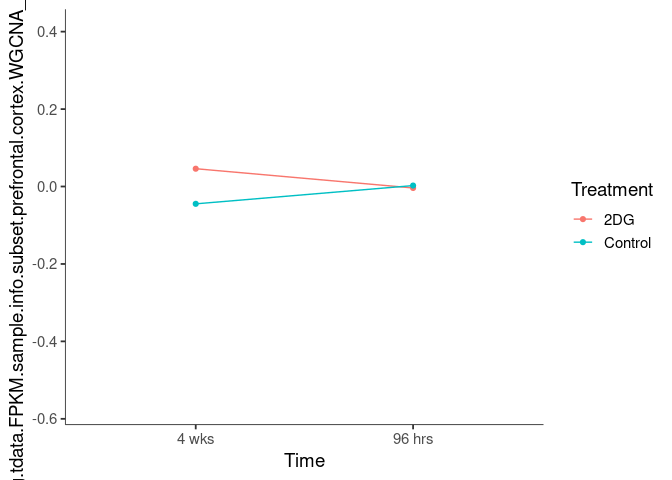
lightskyblue
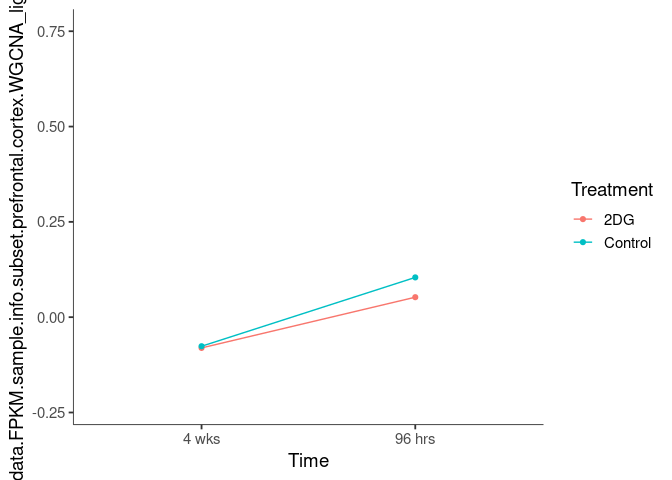
mediumorchid1
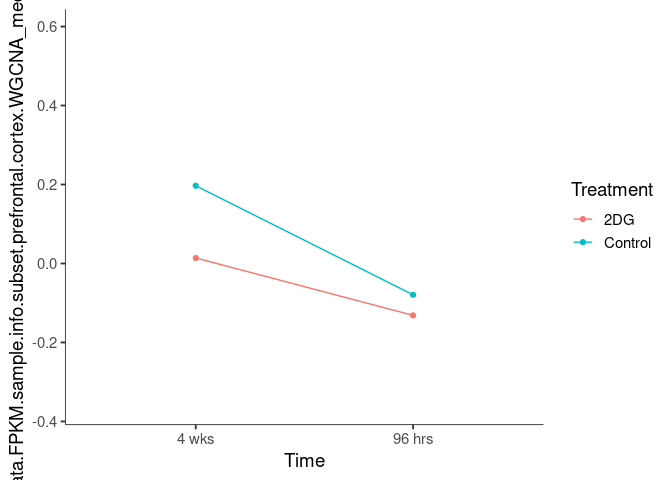
rosybrown2
Analysis performed by Ann Wells
The Carter Lab The Jackson Laboratory 2023
ann.wells@jax.org