Module Sample Contribution Kidney
Ann Wells
April 23, 2023
Introduction and Data files
R Markdown
This dataset contains nine tissues (heart, hippocampus, hypothalamus, kidney, liver, prefrontal cortex, skeletal muscle, small intestine, and spleen) from C57BL/6J mice that were fed 2-deoxyglucose (add dosage) through their drinking water for 96hrs or 4wks. 96hr mice were given their 2DG treatment 2 weeks after the other cohort started the 4 week treatment. The organs from the mice were harvested and processed for metabolomics and transcriptomics. The data in this document pertains to the transcriptomics data only. The counts that were used were FPKM normalized before being log transformed. It was determined that sample A113 had low RNAseq quality and through further analyses with PCA, MA plots, and clustering was an outlier and will be removed for the rest of the analyses performed. This document will determine how samples are contributing to each module.
needed.packages <- c("tidyverse", "here", "functional", "gplots", "dplyr", "GeneOverlap", "R.utils", "reshape2","magrittr","data.table", "RColorBrewer","preprocessCore", "ARTool","emmeans", "phia", "gProfileR","pheatmap")
for(i in 1:length(needed.packages)){library(needed.packages[i], character.only = TRUE)}
source(here("source_files","WGCNA_source.R"))
source(here("source_files","plot_theme.R"))tdata.FPKM.sample.info <- readRDS(here("Data","20190406_RNAseq_B6_4wk_2DG_counts_phenotypes.RData"))
tdata.FPKM <- readRDS(here("Data","20190406_RNAseq_B6_4wk_2DG_counts_numeric.RData"))
log.tdata.FPKM <- log(tdata.FPKM + 1)
log.tdata.FPKM <- as.data.frame(log.tdata.FPKM)
log.tdata.FPKM.sample.info <- cbind(log.tdata.FPKM, tdata.FPKM.sample.info[,27238:27240])
log.tdata.FPKM.sample.info <- log.tdata.FPKM.sample.info %>% rownames_to_column() %>% filter(rowname != "A113") %>% column_to_rownames()
log.tdata.FPKM.subset <- log.tdata.FPKM[,colMeans(log.tdata.FPKM != 0) > 0.5]
log.tdata.FPKM.sample.info.subset <- cbind(log.tdata.FPKM.subset,tdata.FPKM.sample.info[,27238:27240])
log.tdata.FPKM.sample.info.subset <- log.tdata.FPKM.sample.info.subset %>% rownames_to_column() %>% filter(rowname != "A113") %>% column_to_rownames()
log.tdata.FPKM.sample.info.subset.kidney <- log.tdata.FPKM.sample.info.subset %>% rownames_to_column() %>% filter(Tissue == "Kidney") %>% column_to_rownames()
modules <- read.csv(here("Data","Kidney","log.tdata.FPKM.sample.info.subset.kidney.WGCNA.module.membership.csv"), header=T)
eigens <- read.csv(here("Data","Kidney","log.tdata.FPKM.sample.info.subset.kidney.WGCNA.module.eigens.csv"), header=T)Module Sample Contribution
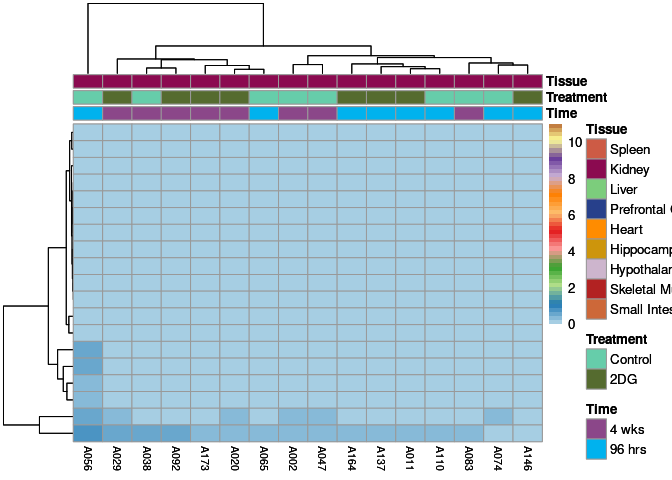
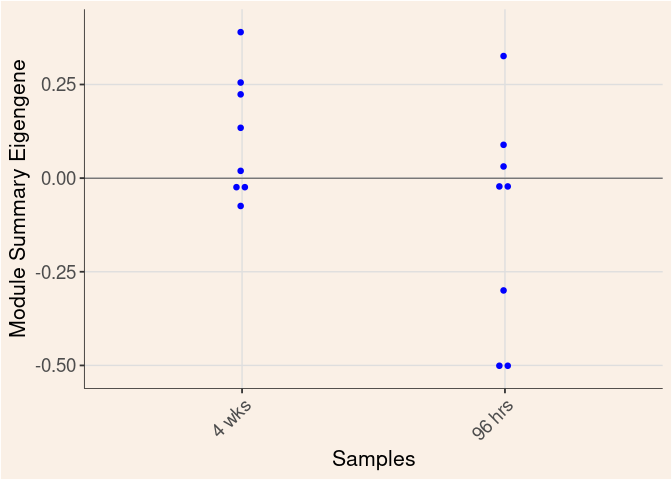
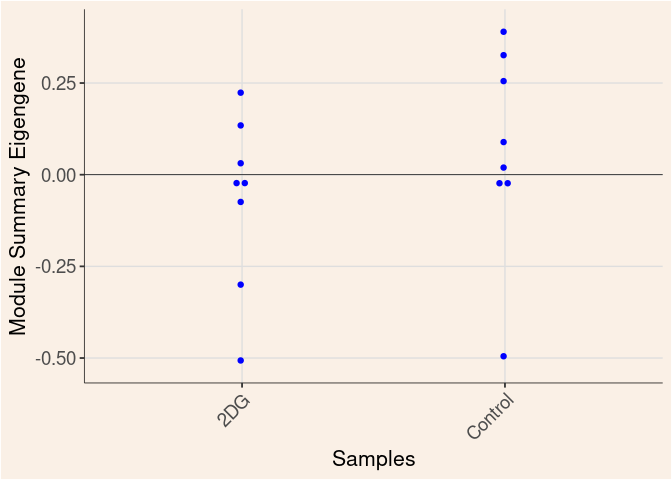
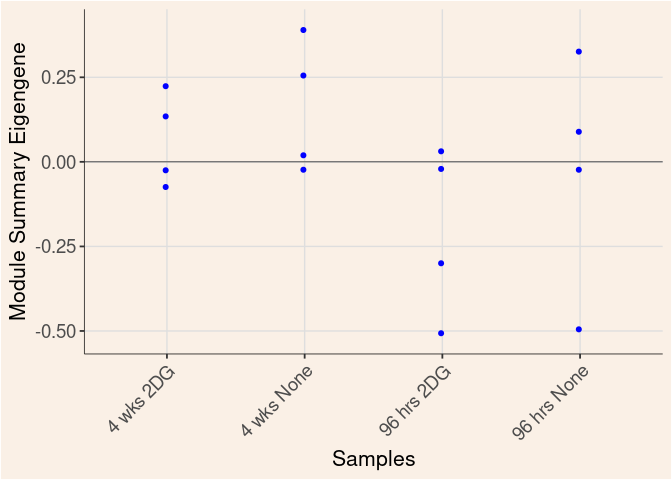
The heatmap shows the expression level of each gene in the module across all samples present in this subsetted dataset. The bar plot shows the relative eigen value summarizing gene expression for each sample present in this subsetted dataset.
eigen.expression(eigens,log.tdata.FPKM.sample.info.subset.kidney)cyan Module
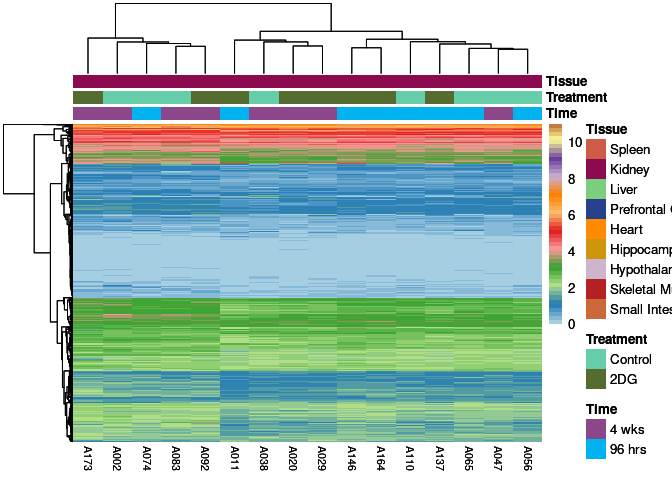
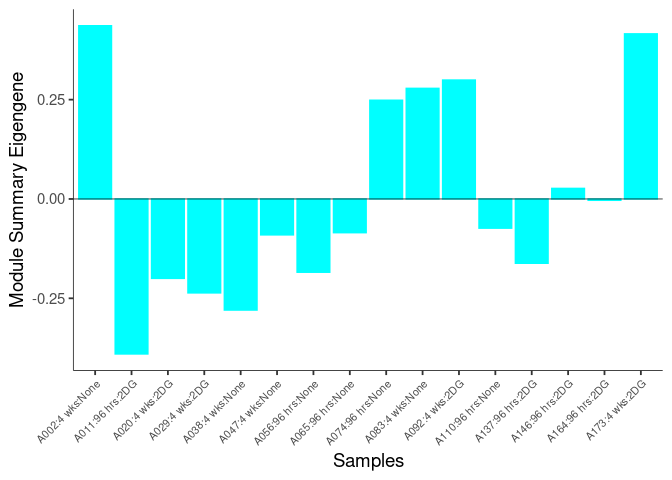
blue Module
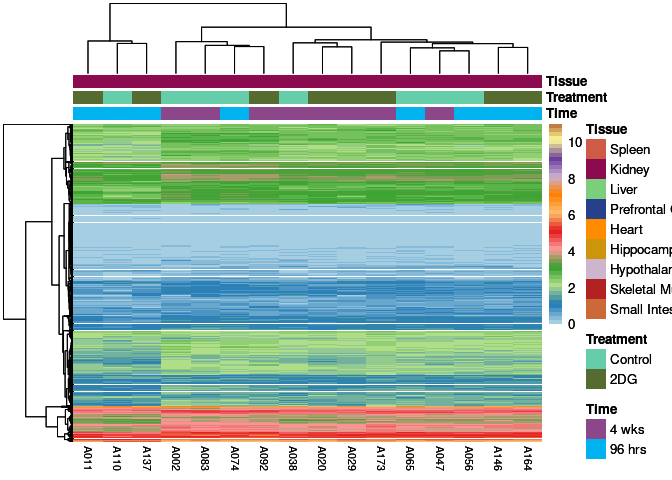
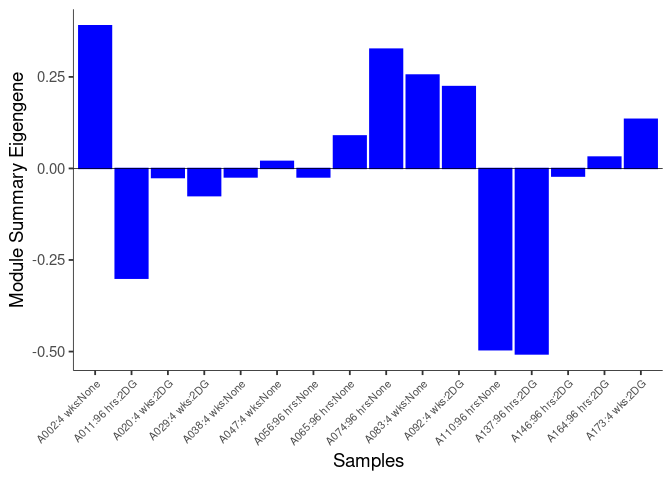
red Module
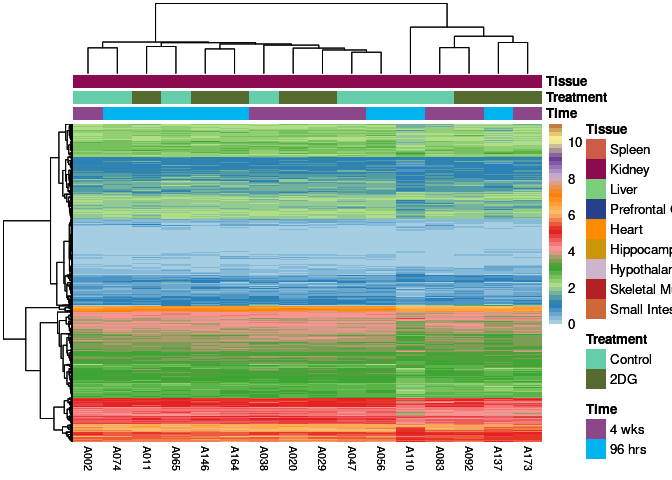
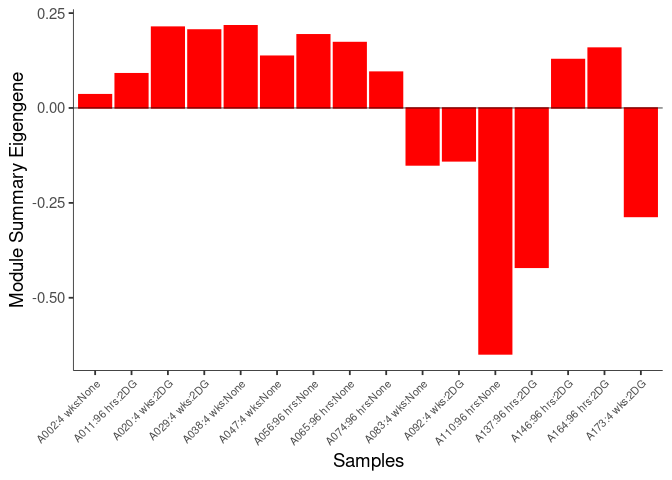
pink Module
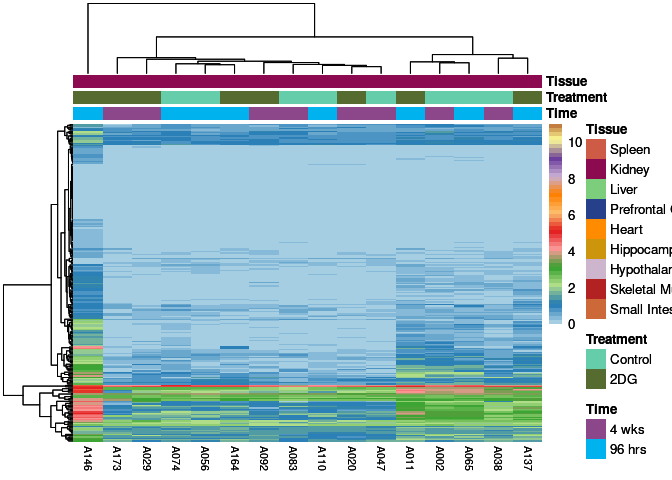
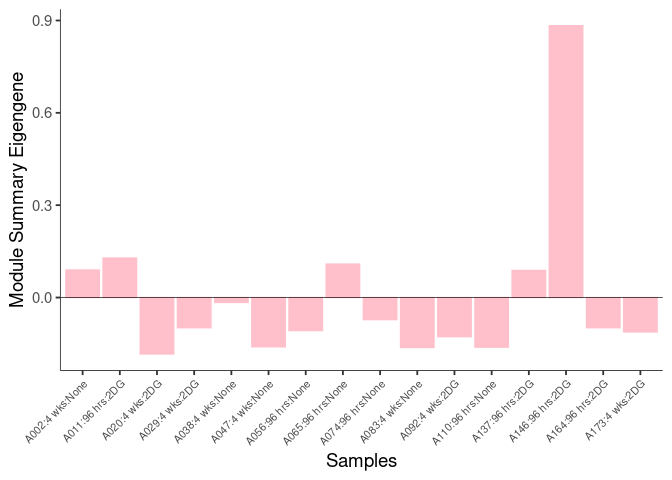
magenta Module
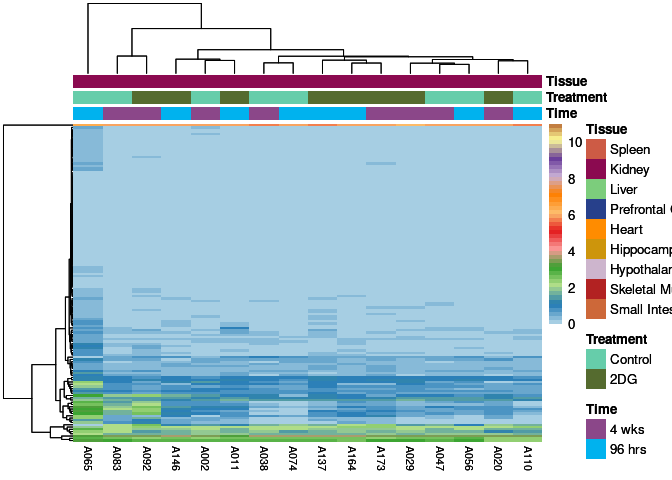
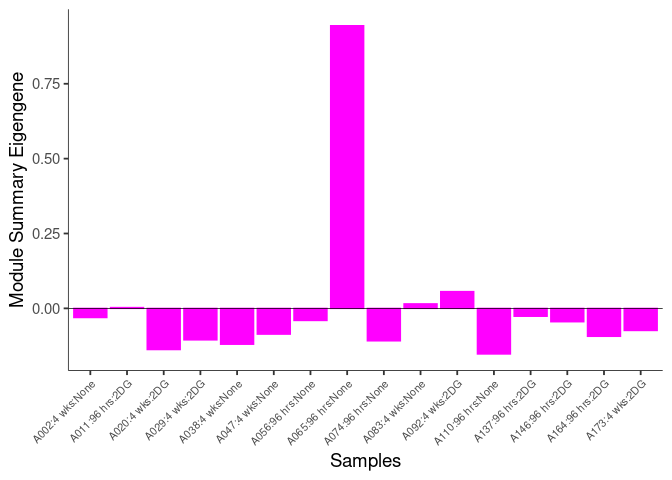
darkred Module
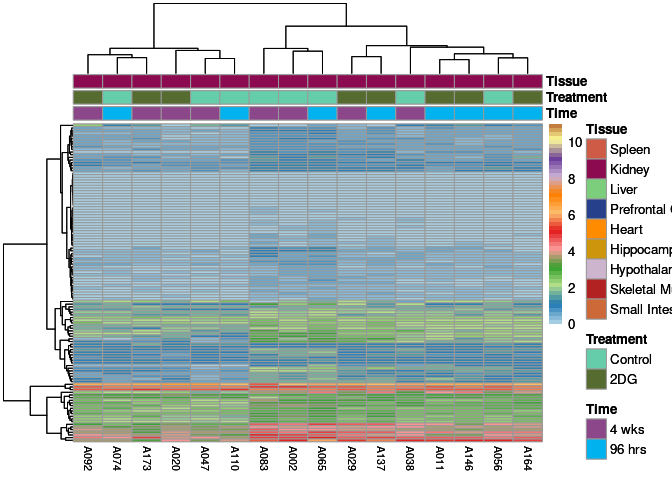
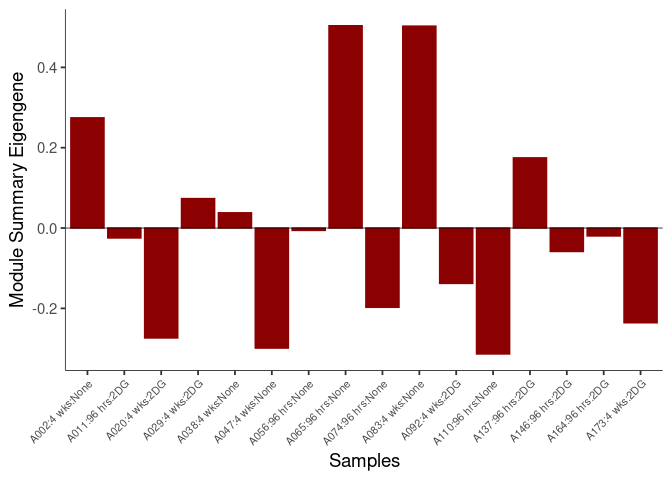
midnightblue Module
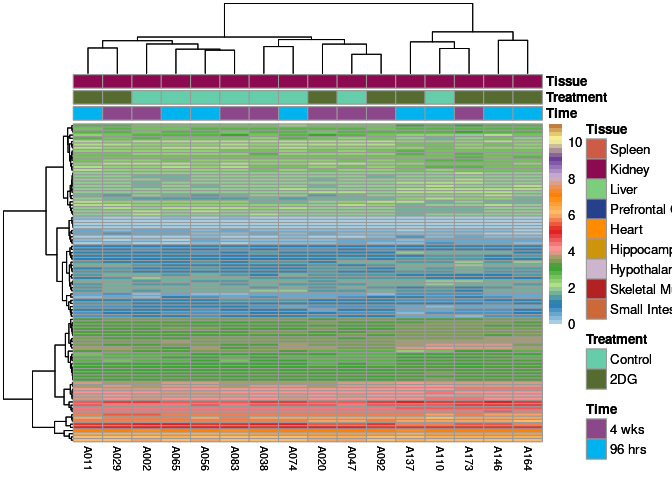
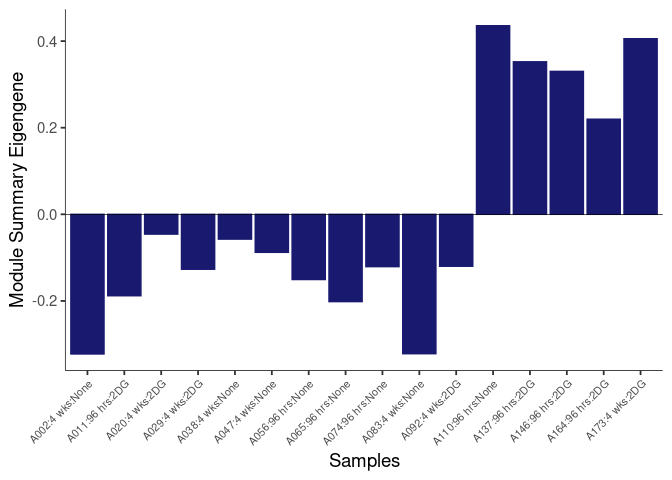
purple Module
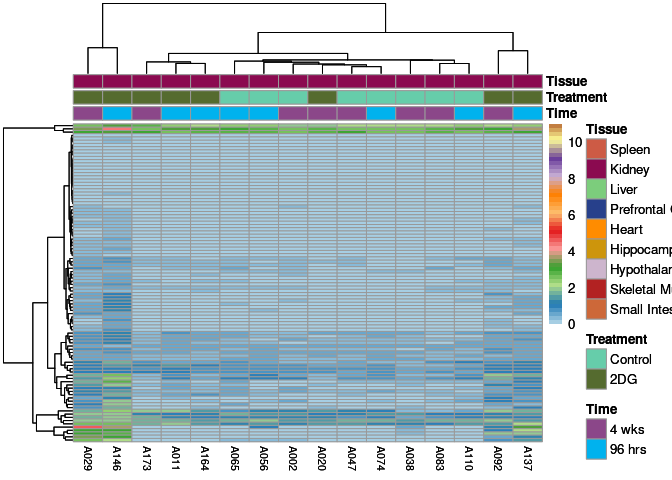
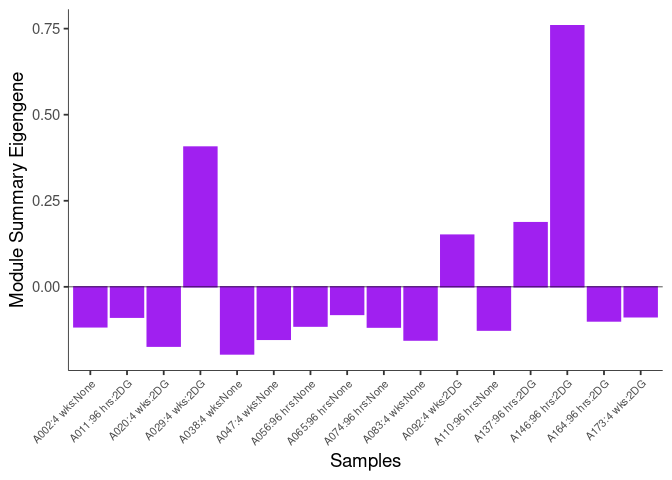
greenyellow Module
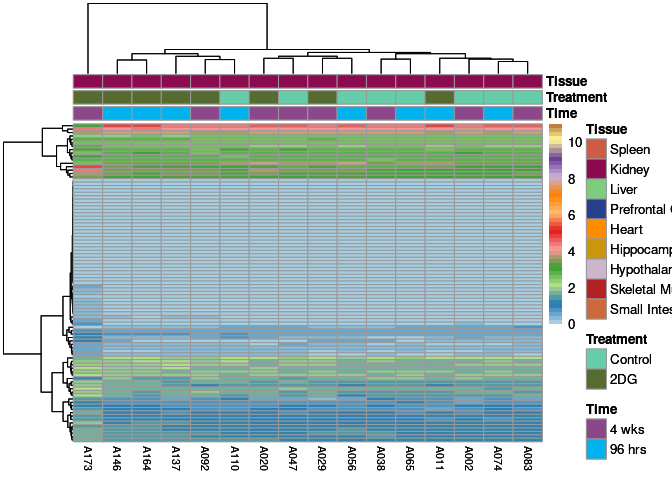
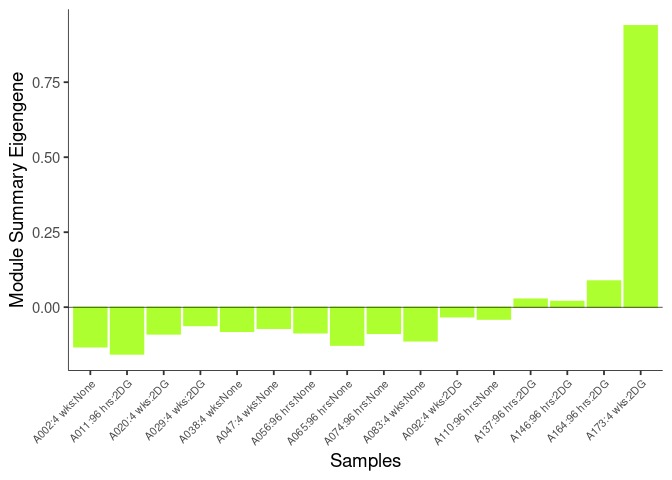
salmon Module
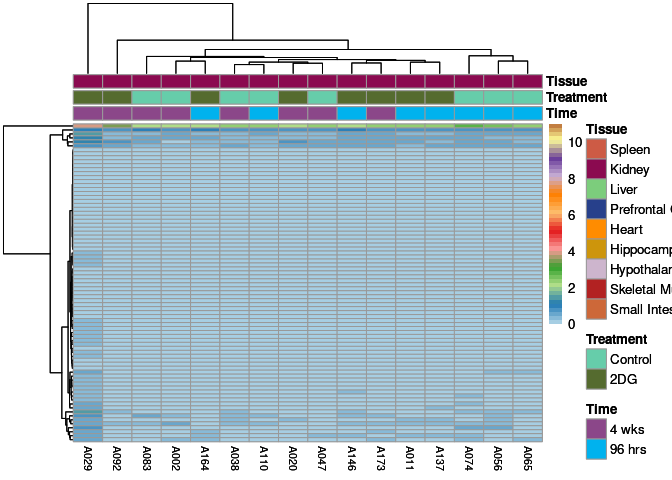
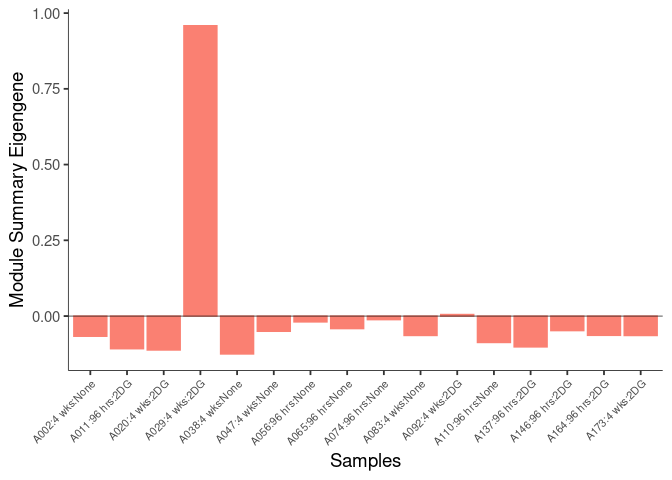
lightcyan Module
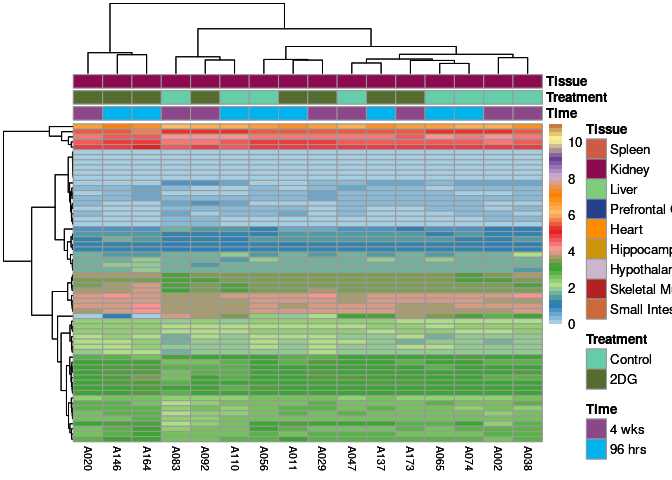
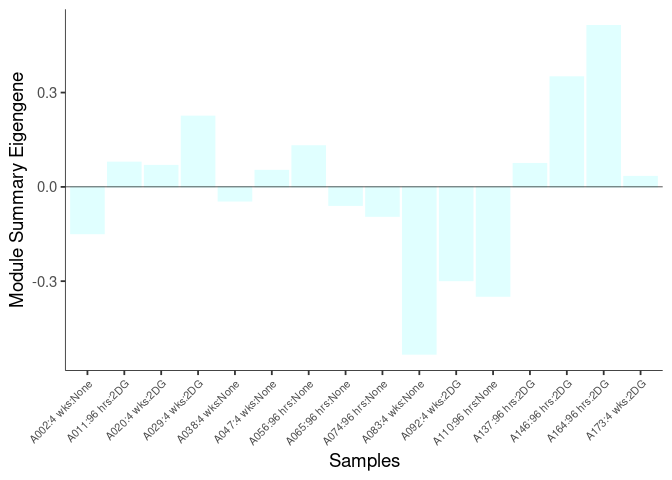
grey60 Module
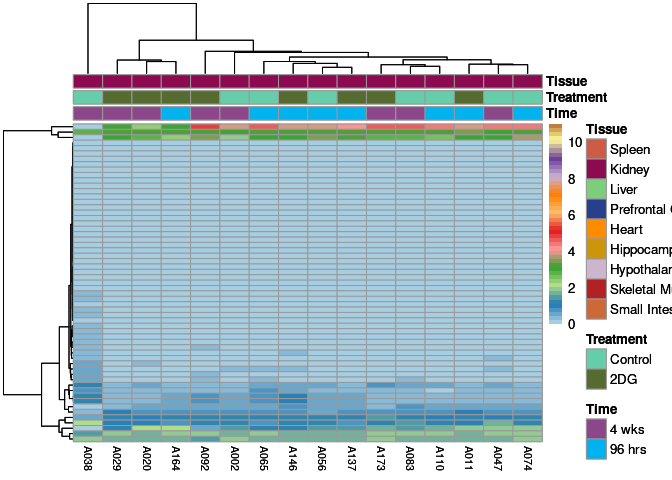
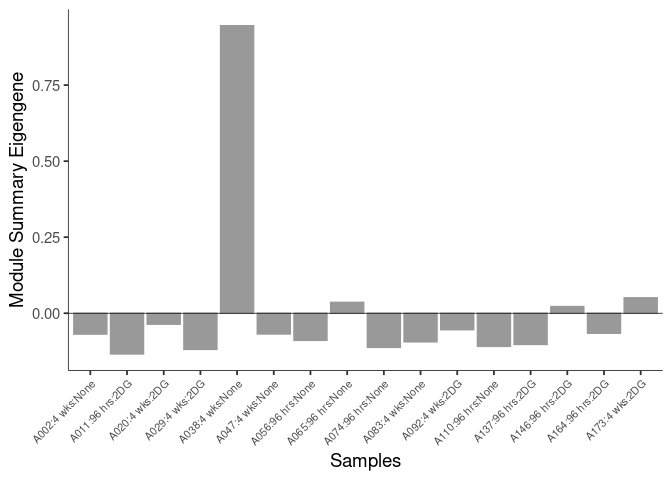
lightgreen Module
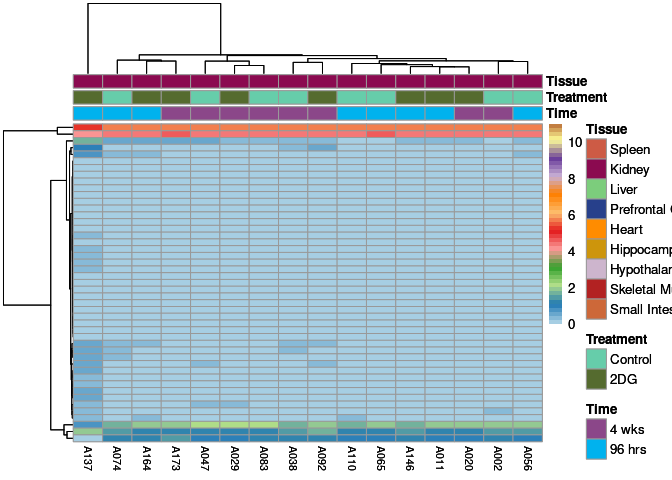
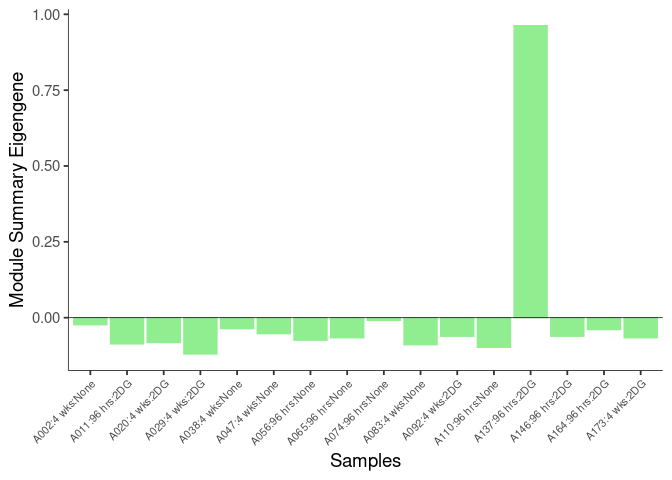
lightyellow Module
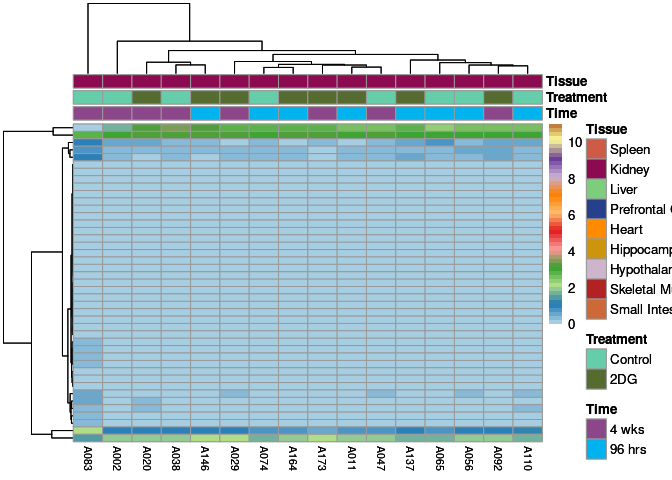
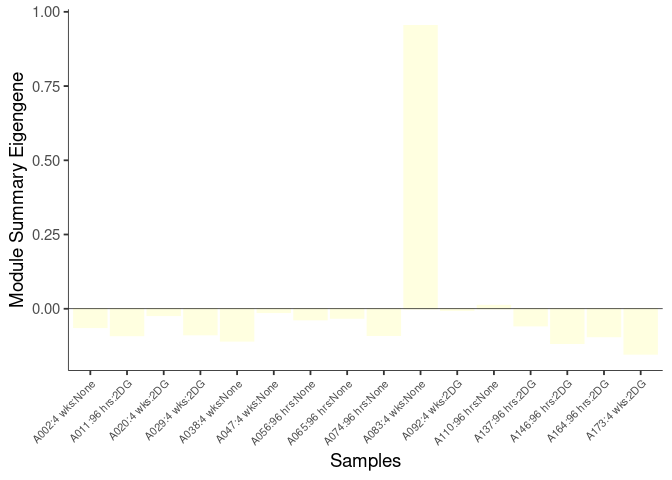
darkgreen Module
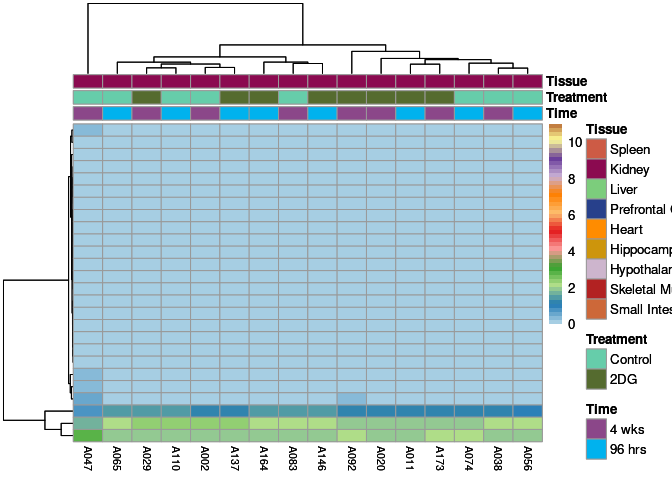
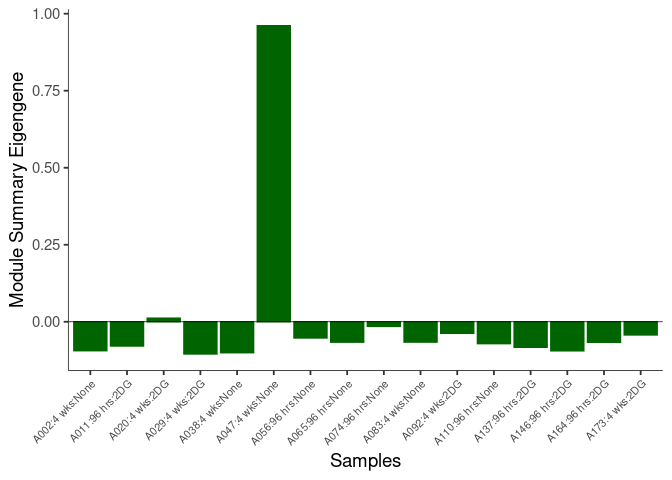
darkturquoise Module
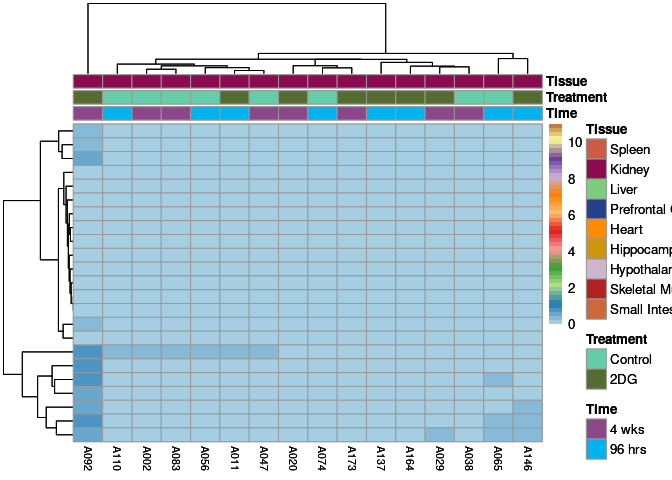
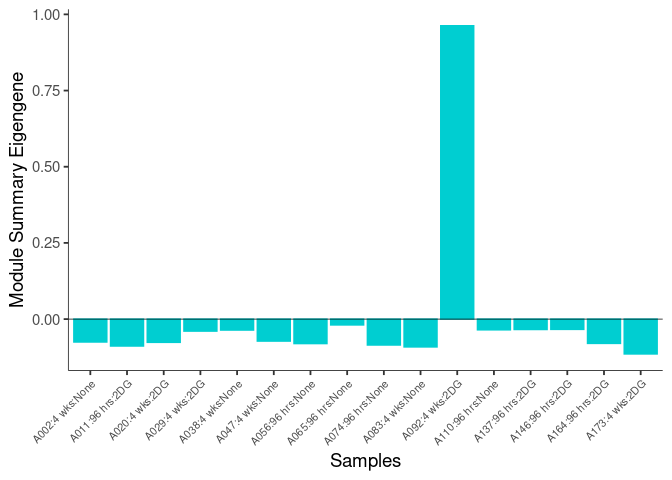
darkgrey Module
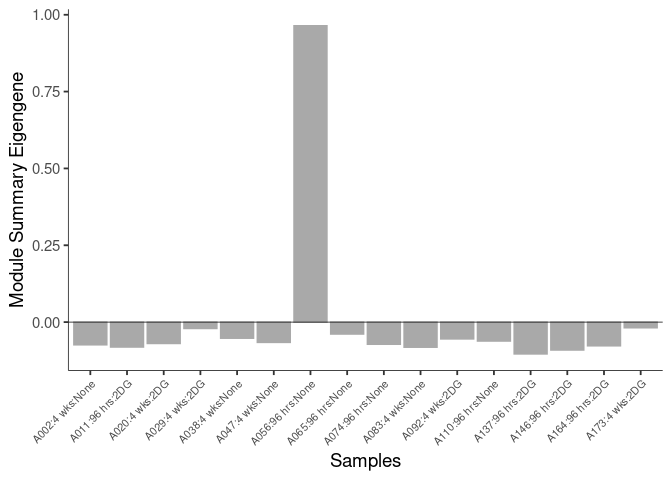
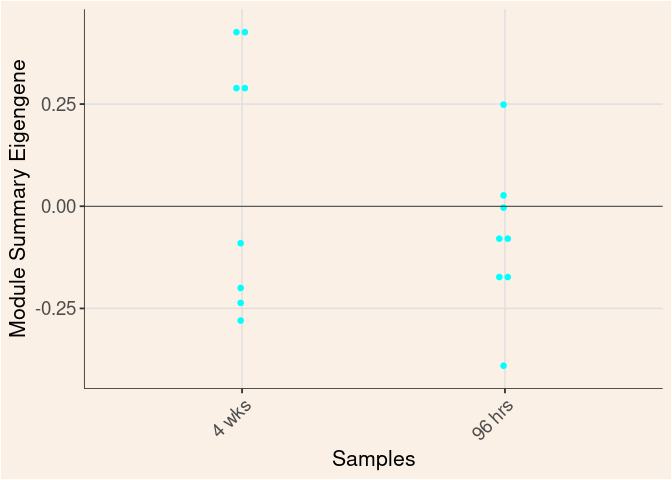
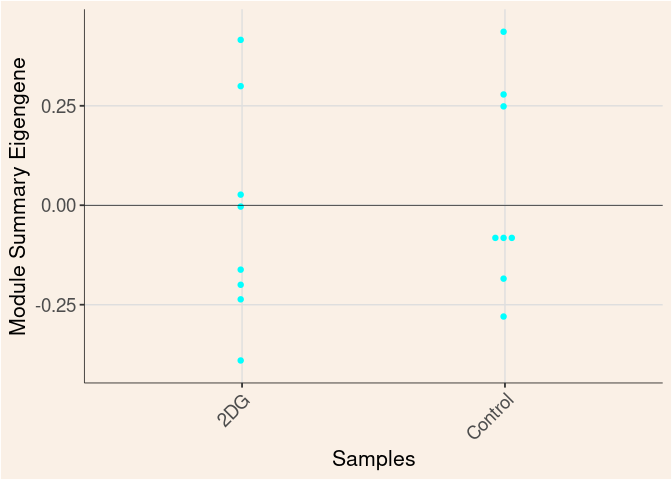
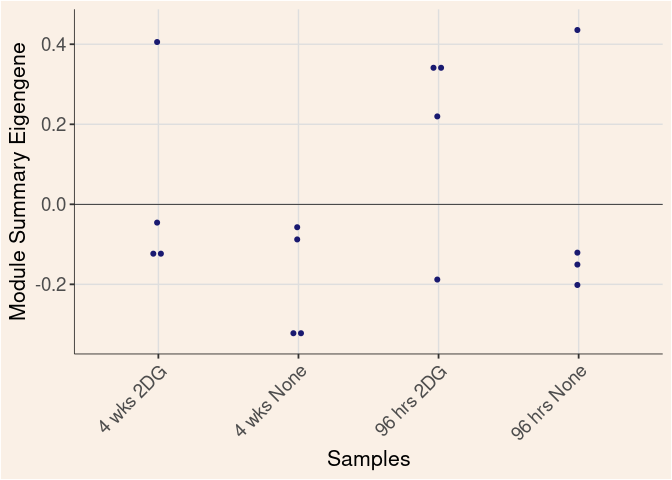
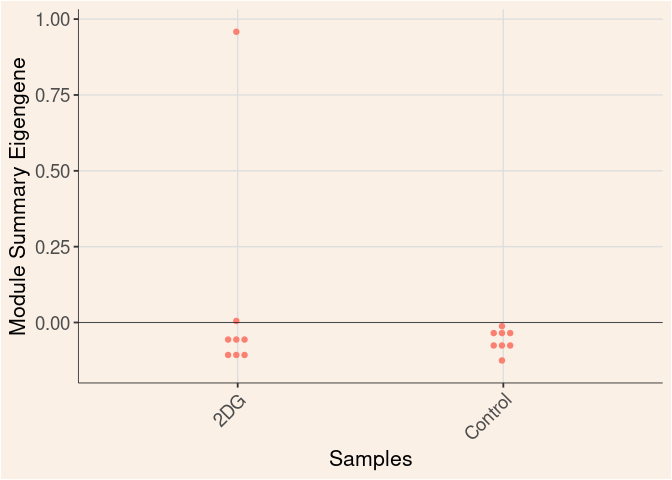
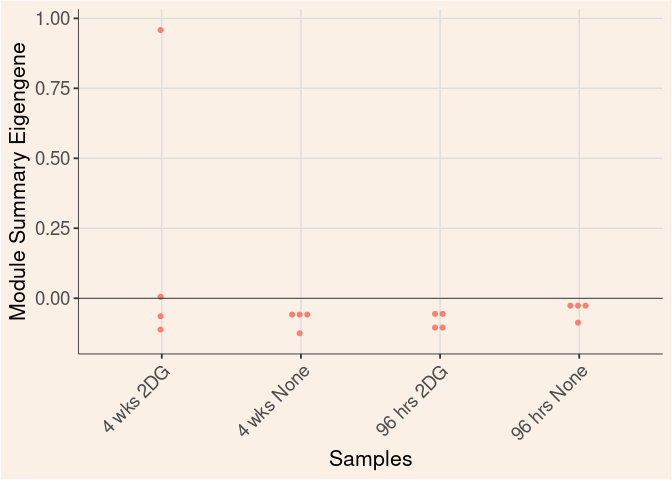
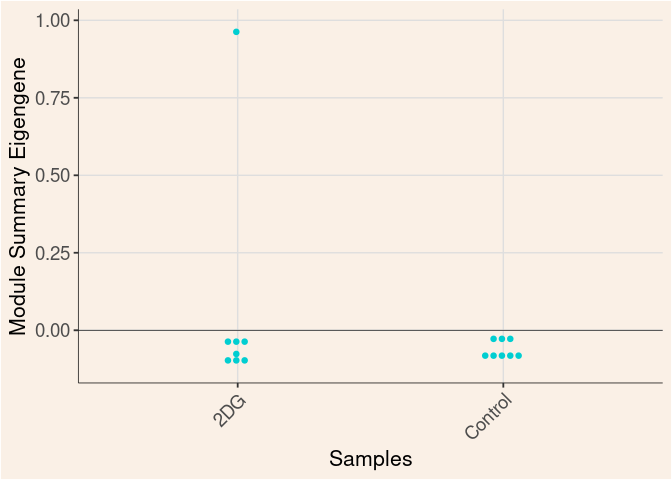
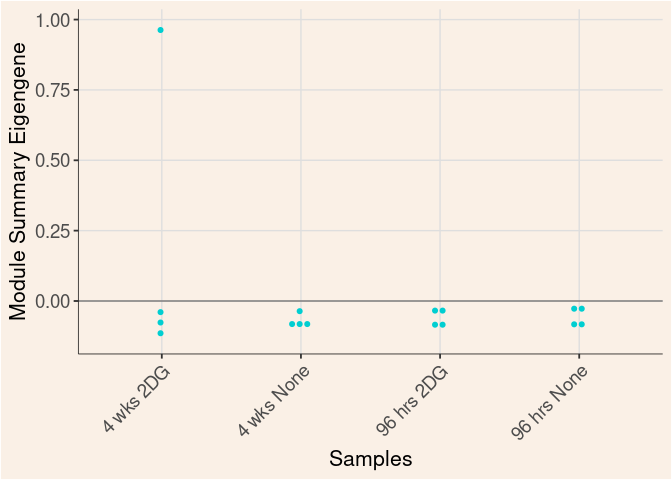
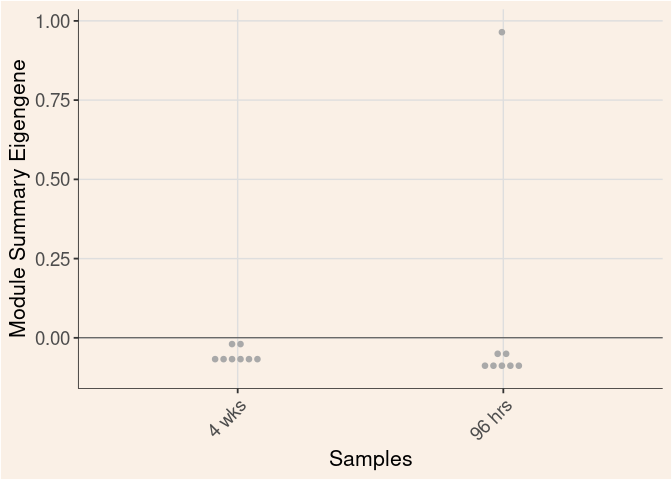
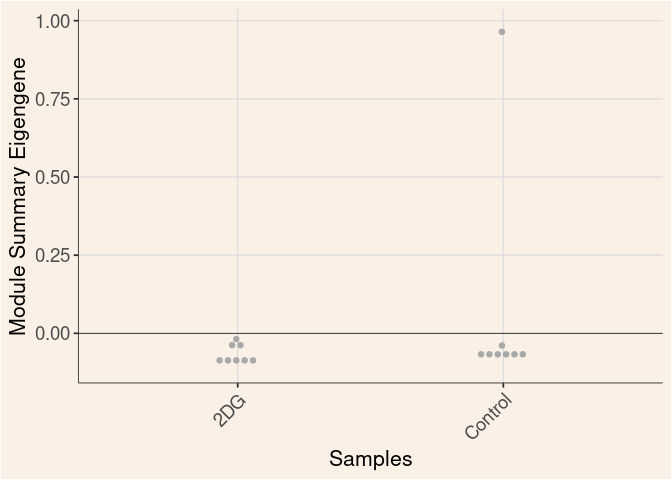
Module Sample Contribution Dot plot
dot.plot(eigens)cyan Module
Time
Treatment
Time_by_Treatment
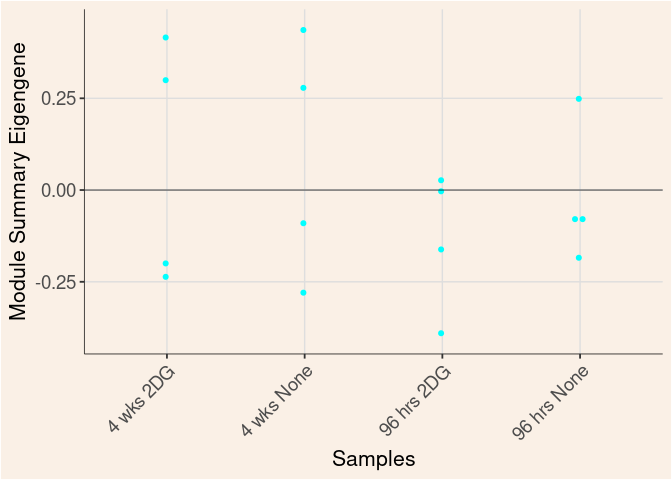
blue Module
Time
Treatment
Time_by_Treatment
red Module
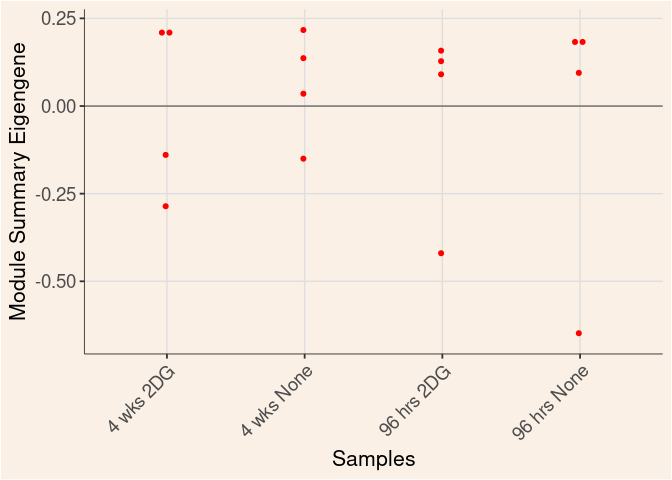
Time
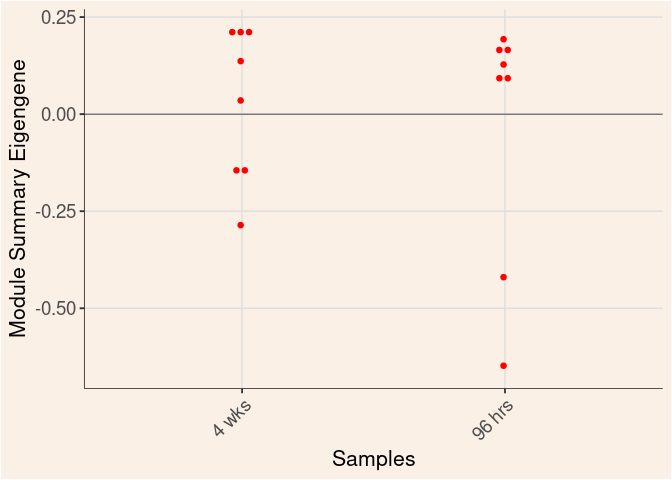
Treatment
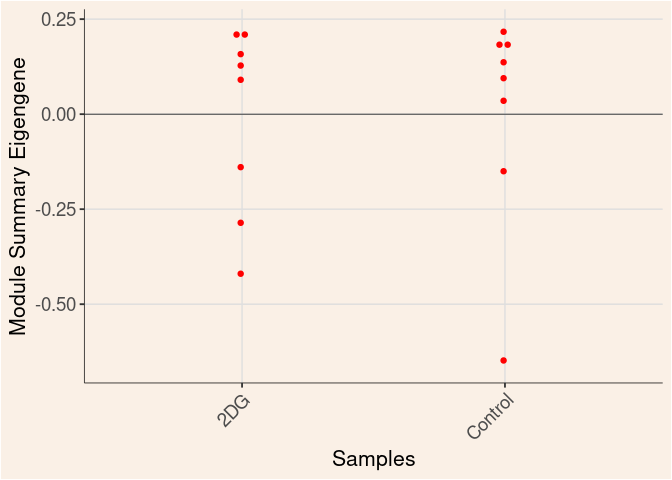
Time_by_Treatment
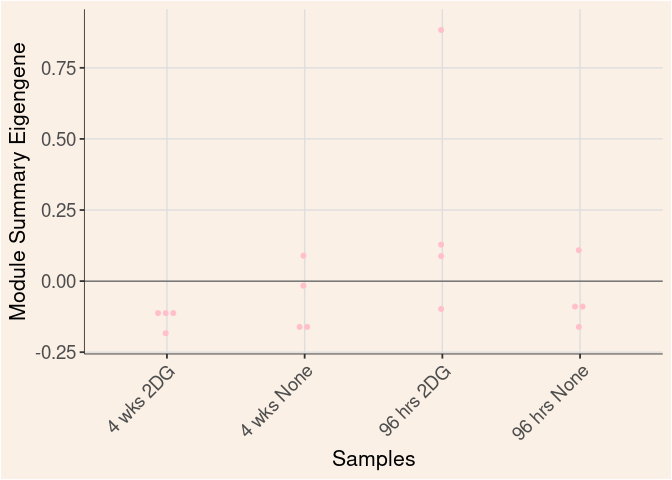
pink Module
Time
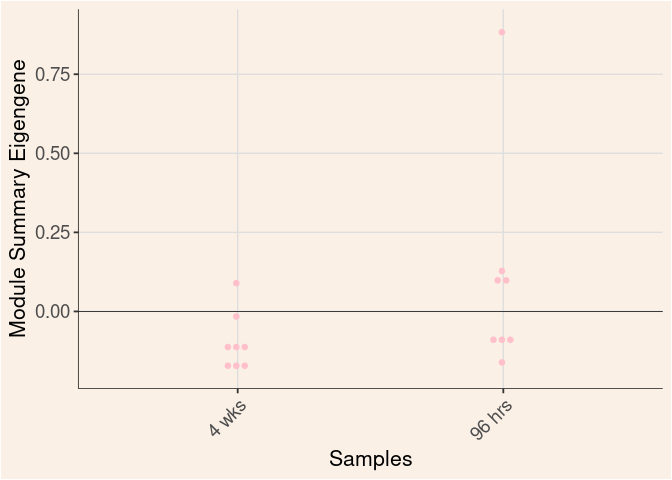
Treatment
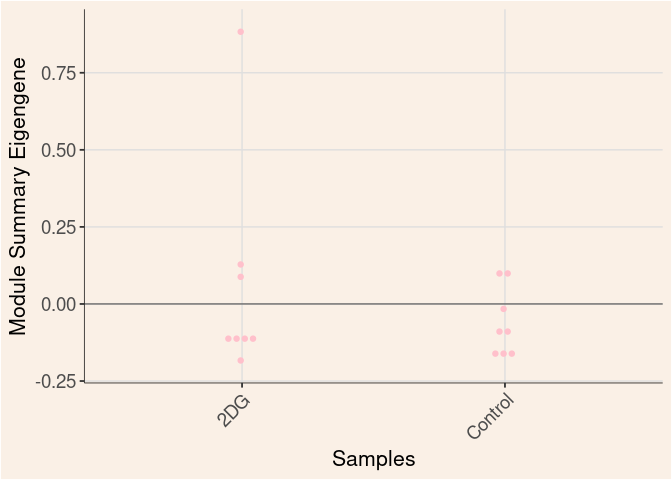
Time_by_Treatment
magenta Module
Time
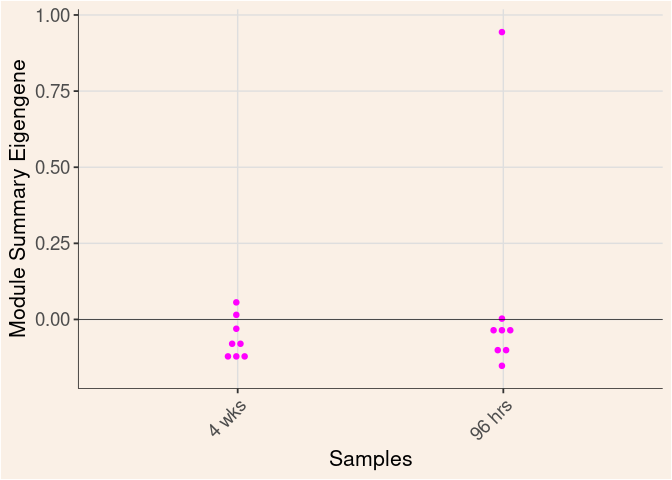
Treatment
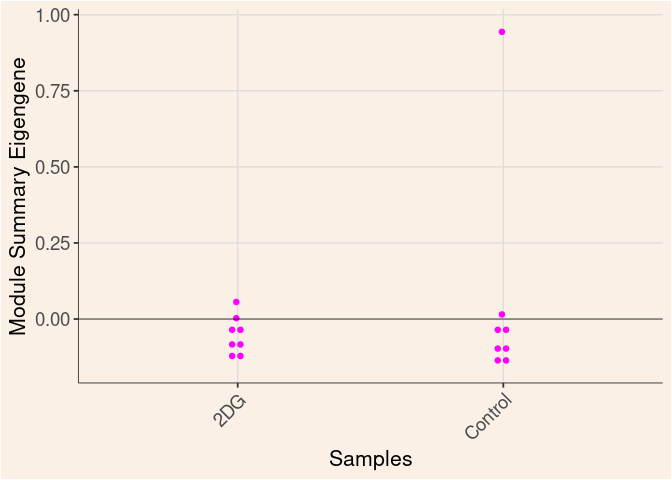
Time_by_Treatment
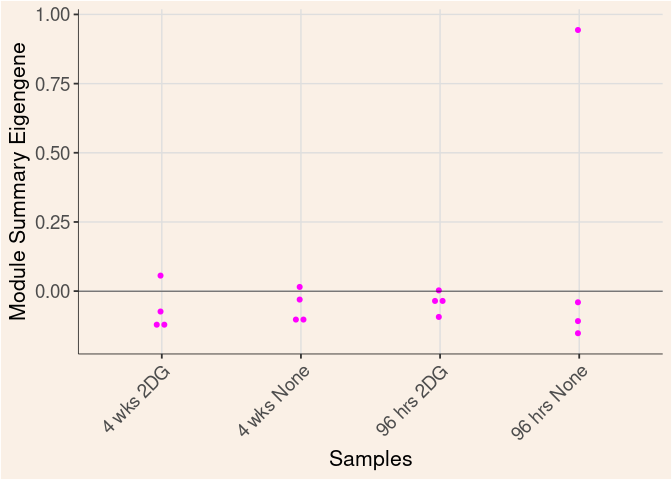
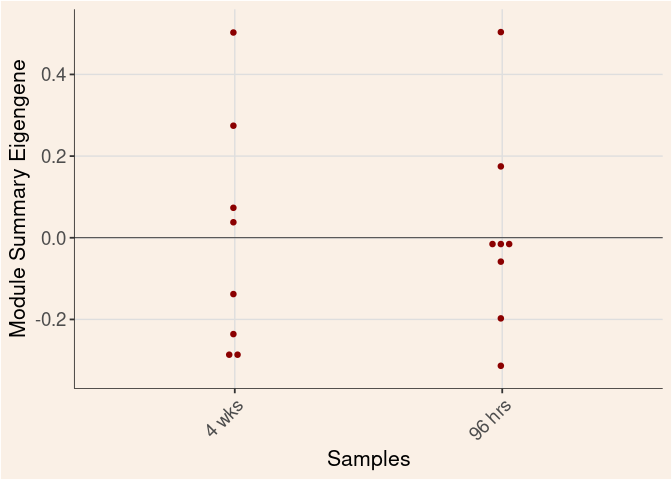
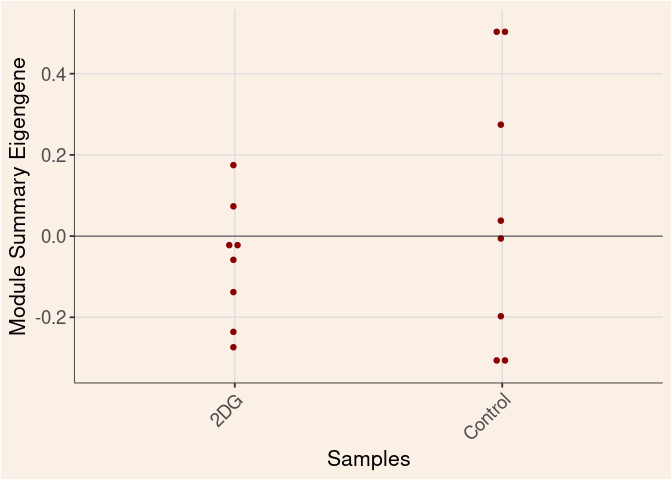
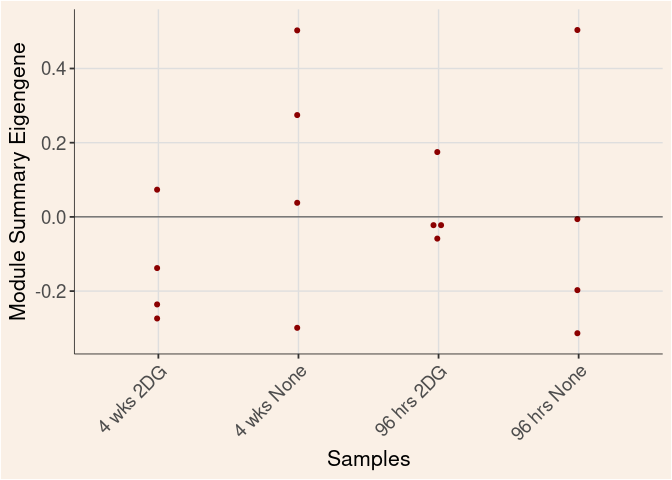
darkred Module
Time
Treatment
Time_by_Treatment
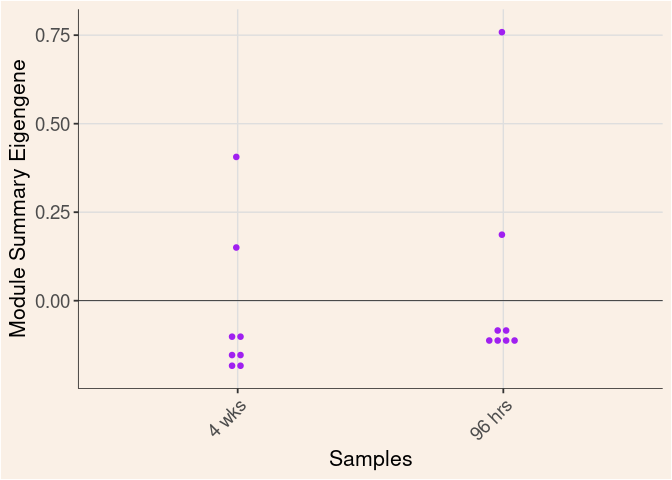
midnightblue Module
Time
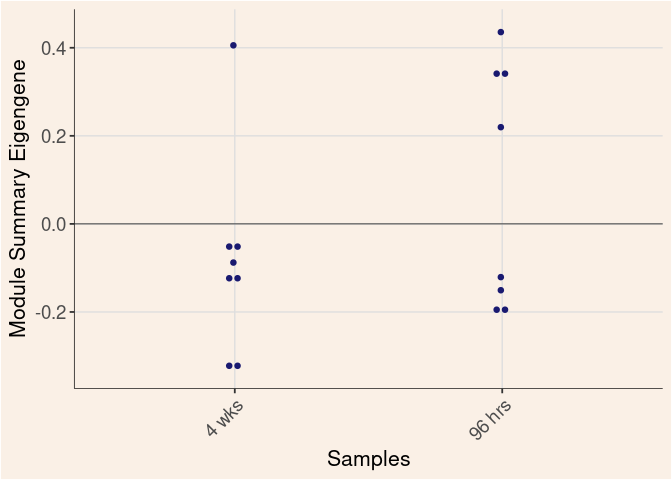
Treatment
Time_by_Treatment
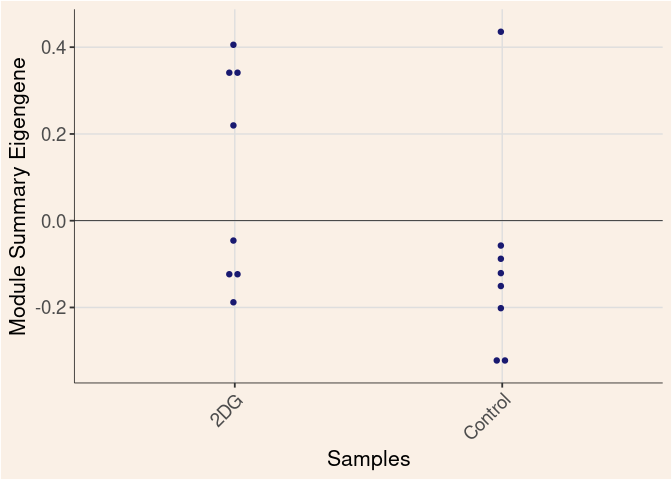
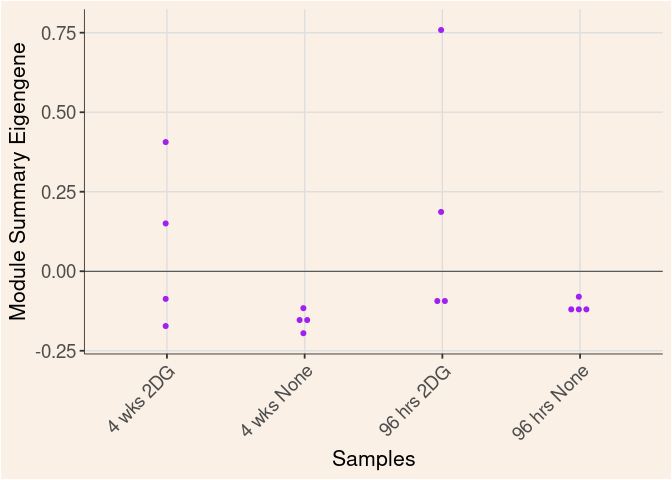
purple Module
Time
Treatment
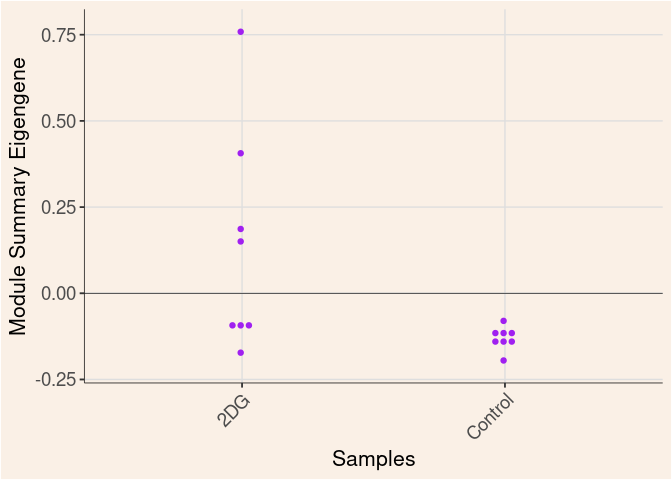
Time_by_Treatment
greenyellow Module
Time
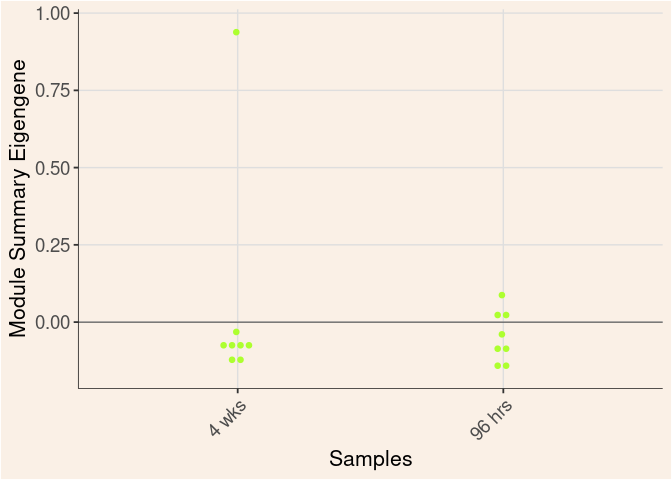
Treatment
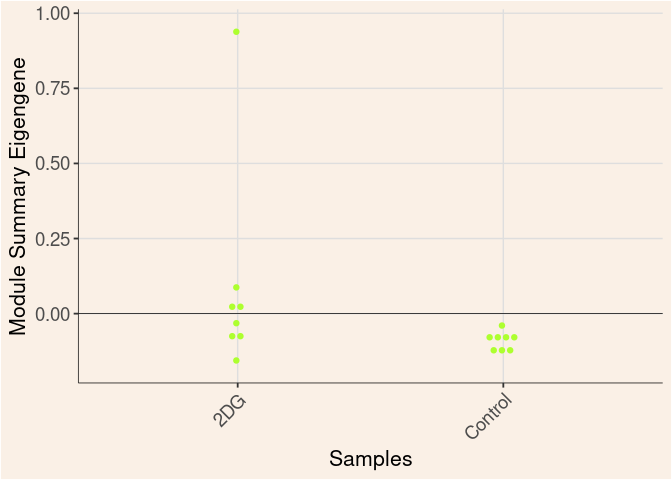
Time_by_Treatment
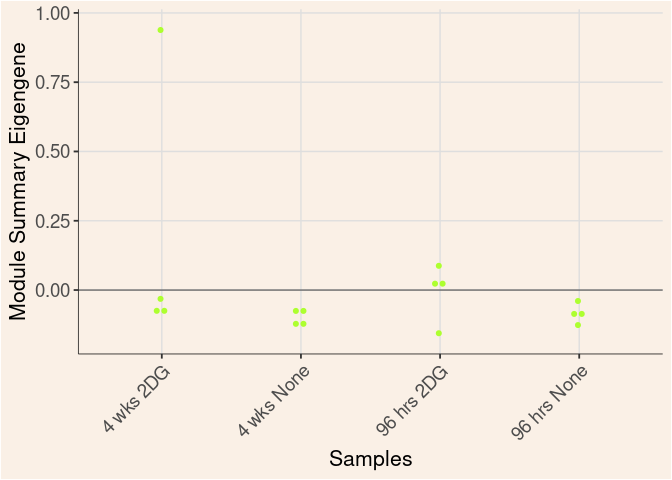
salmon Module
Time
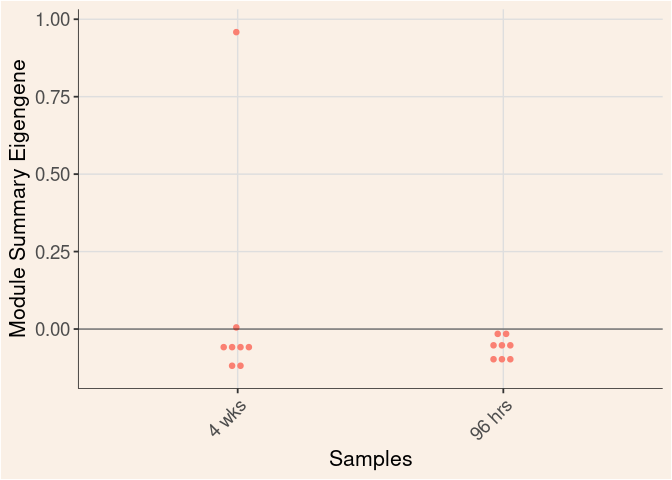
Treatment
Time_by_Treatment
lightcyan Module
Time
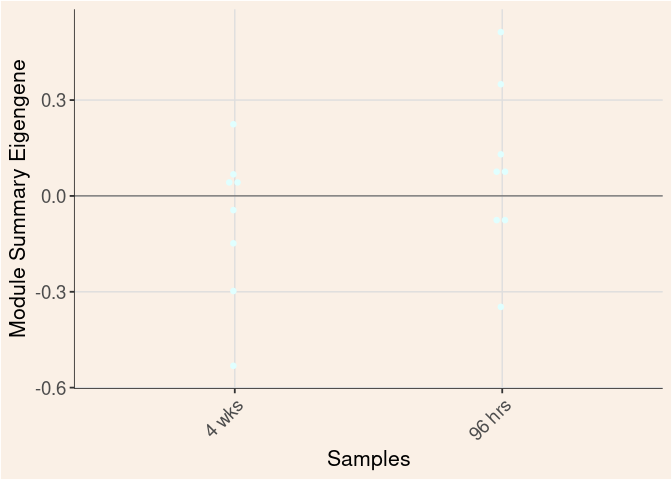
Treatment
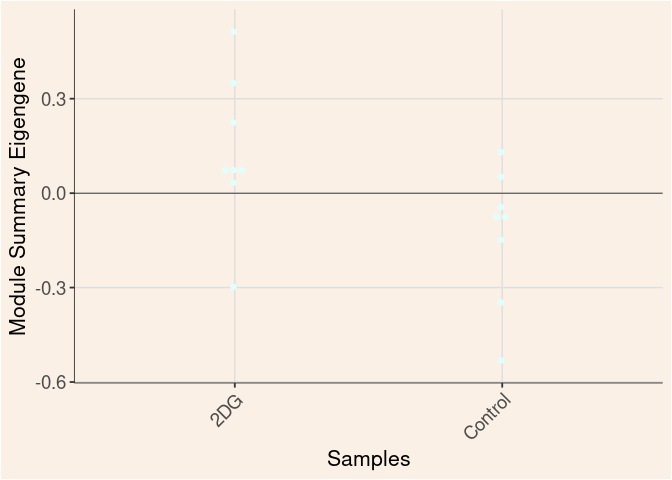
Time_by_Treatment
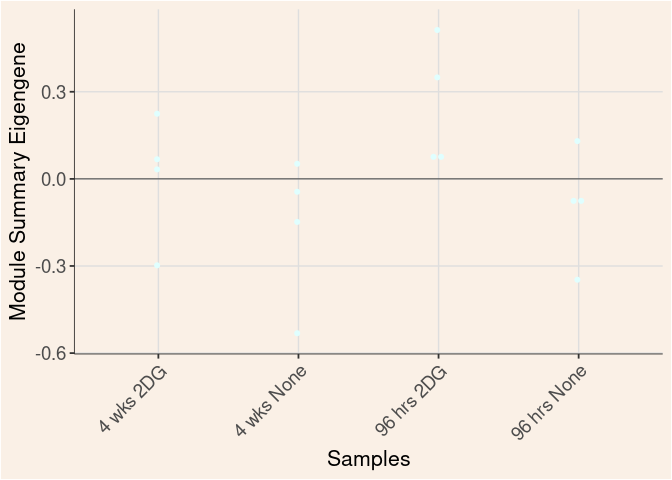
grey60 Module
Time
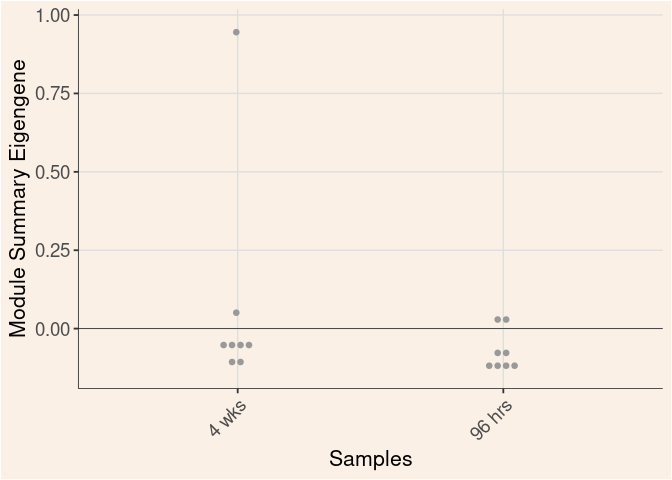
Treatment
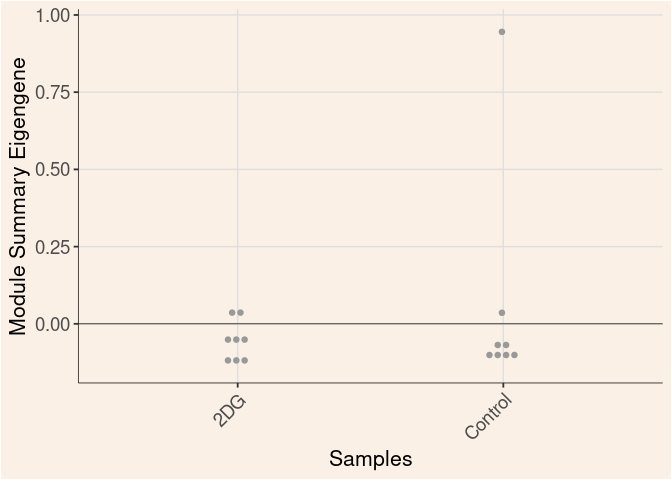
Time_by_Treatment
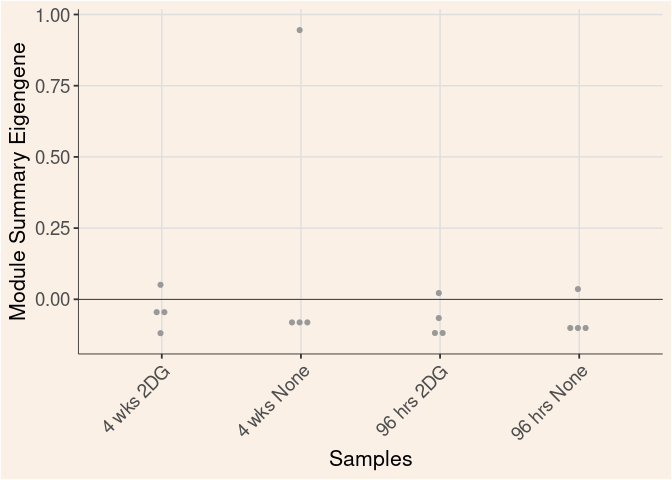
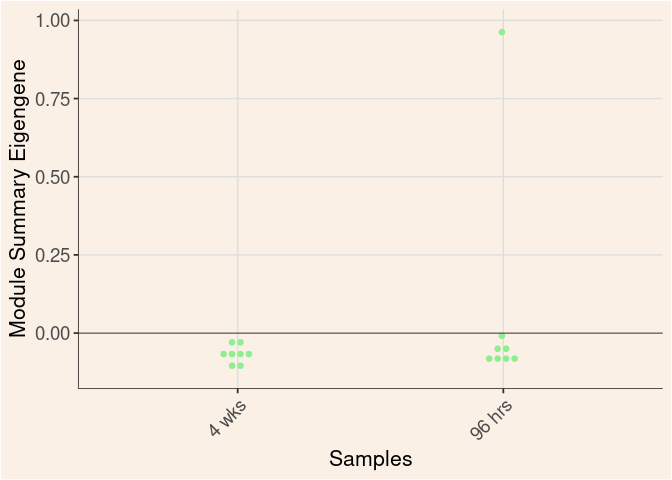
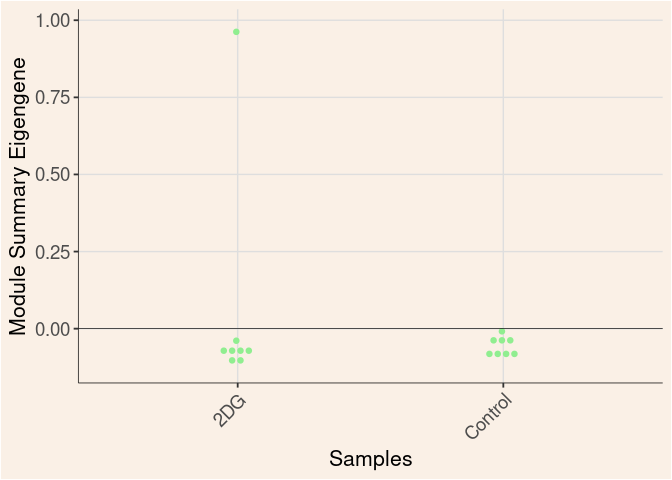
lightgreen Module
Time
Treatment
Time_by_Treatment
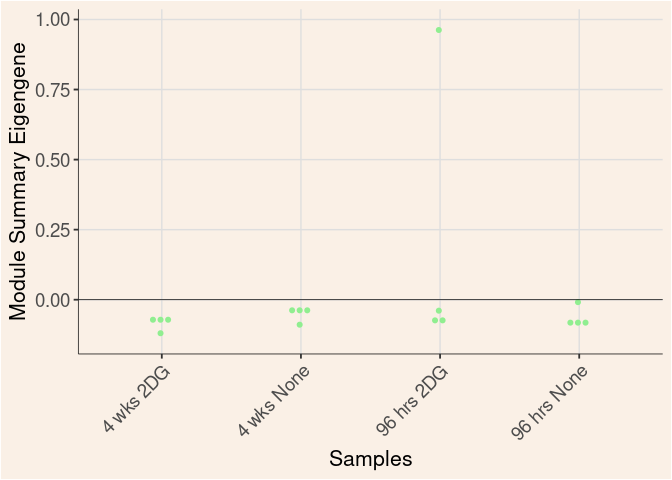
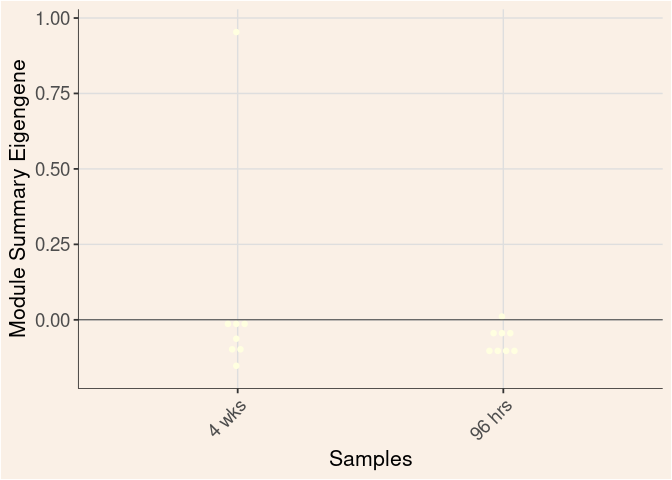
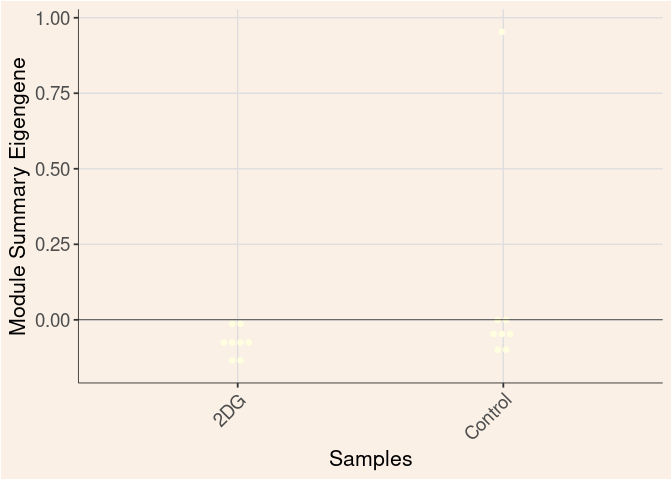
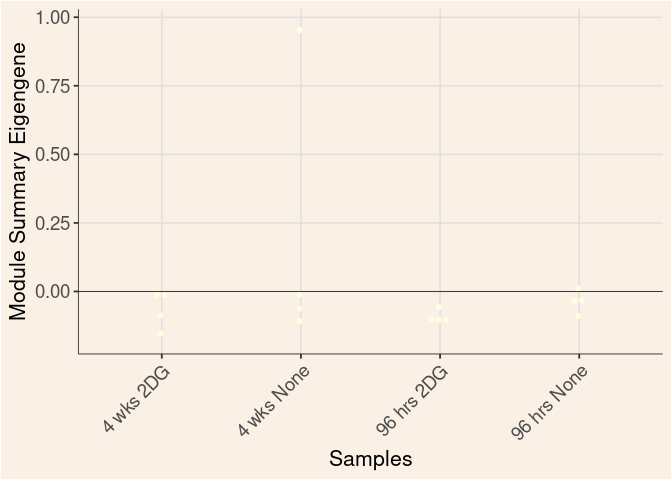
lightyellow Module
Time
Treatment
Time_by_Treatment
darkgreen Module
Time
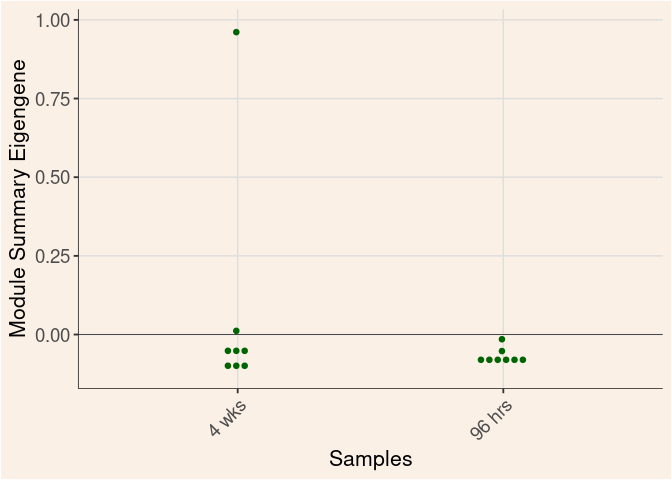
Treatment
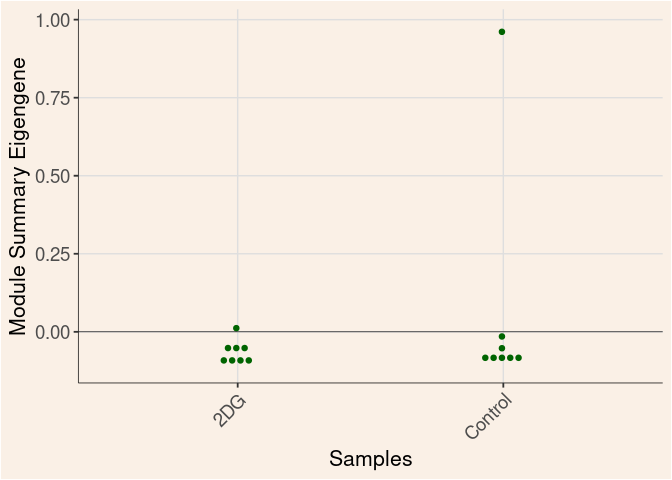
Time_by_Treatment
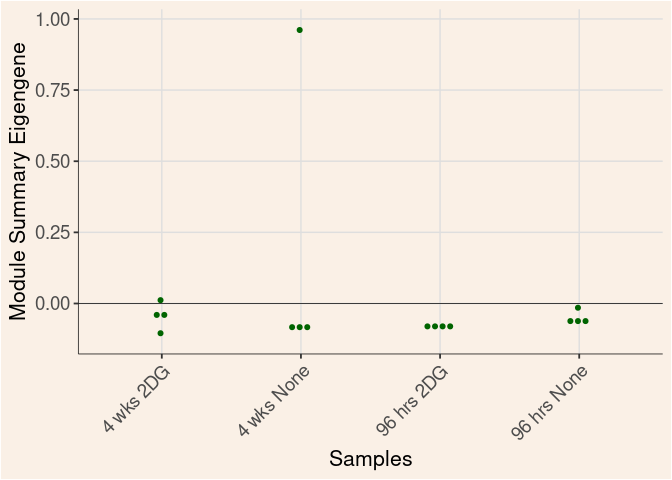
darkturquoise Module
Time
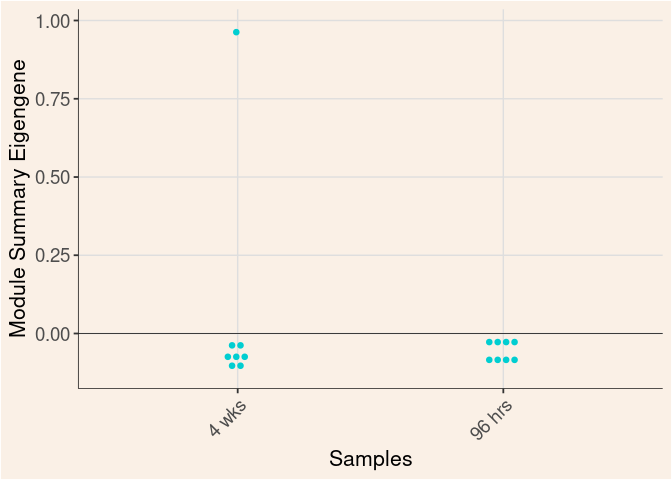
Treatment
Time_by_Treatment
darkgrey Module
Time
Treatment
Time_by_Treatment
Analysis performed by Ann Wells
The Carter Lab The Jackson Laboratory 2023
ann.wells@jax.org