Data Cleaning Prefrontal Cortex
Ann Wells
April 23, 2023
tdata.FPKM.sample.info <- readRDS(here("Data","20190406_RNAseq_B6_4wk_2DG_counts_phenotypes.RData"))
tdata.FPKM <- readRDS(here("Data","20190406_RNAseq_B6_4wk_2DG_counts_numeric.RData"))
log.tdata.FPKM <- log(tdata.FPKM + 1)
log.tdata.FPKM <- as.data.frame(log.tdata.FPKM)
log.tdata.FPKM.sample.info <- cbind(log.tdata.FPKM, tdata.FPKM.sample.info[,27238:27240])
log.tdata.FPKM.sample.info <- log.tdata.FPKM.sample.info %>% rownames_to_column() %>% filter(rowname != "A113") %>% column_to_rownames()
log.tdata.FPKM.subset <- log.tdata.FPKM[,colMeans(log.tdata.FPKM != 0) > 0.5]
log.tdata.FPKM.sample.info.subset <- cbind(log.tdata.FPKM.subset,tdata.FPKM.sample.info[,27238:27240])
log.tdata.FPKM.sample.info.subset <- log.tdata.FPKM.sample.info.subset %>% rownames_to_column() %>% filter(rowname != "A113") %>% column_to_rownames()
log.tdata.FPKM.sample.info.subset.prefrontal.cortex <- log.tdata.FPKM.sample.info.subset %>% rownames_to_column() %>% filter(Tissue == "Pre-frontal Cortex") %>% column_to_rownames()Wrangle Data
I will use Mouse IDs Treatment, and Time to keep track of the values in the matrices. All other covariates will be discarded.
# Set rownames by mouse ID and tissue
rownames(log.tdata.FPKM.sample.info.subset.prefrontal.cortex) <- paste0(rownames(log.tdata.FPKM.sample.info.subset.prefrontal.cortex),":", log.tdata.FPKM.sample.info.subset.prefrontal.cortex$Time, ":", log.tdata.FPKM.sample.info.subset.prefrontal.cortex$Treatment)
# Discard covariates from columns 17333-17336
log.tdata.FPKM.sample.info.subset.prefrontal.cortex <- log.tdata.FPKM.sample.info.subset.prefrontal.cortex[,-(17333:17336)]
head(log.tdata.FPKM.sample.info.subset.prefrontal.cortex[,1:5])## ENSMUSG00000000001 ENSMUSG00000000028 ENSMUSG00000000031
## A009:4 wks:None 2.636912 0.6259384 0.1655144
## A018:96 hrs:2DG 3.065258 0.7129498 0.1310283
## A027:4 wks:2DG 2.755570 0.6626880 0.1043600
## A036:4 wks:2DG 2.702703 0.7419373 0.0861777
## A045:4 wks:None 2.451867 0.4574248 0.7275486
## A054:4 wks:None 2.588516 0.7419373 0.3148107
## ENSMUSG00000000037 ENSMUSG00000000049
## A009:4 wks:None 0.2700271 0.6097656
## A018:96 hrs:2DG 0.4121097 0.8372475
## A027:4 wks:2DG 0.8671005 0.4317824
## A036:4 wks:2DG 0.2623643 0.4637340
## A045:4 wks:None 0.4824261 0.3293037
## A054:4 wks:None 0.2390169 0.7178398Check Data for Missing Values
WGCNA will have poor results if the data have too many missing values. I checked if any metabolites fall into this category.
log.tdata.FPKM.sample.info.subset.prefrontal.cortex.missing <- missing(log.tdata.FPKM.sample.info.subset.prefrontal.cortex)
cat("logFPKM: ", goodSamplesGenes(log.tdata.FPKM.sample.info.subset.prefrontal.cortex.missing, verbose=0)$allOK, "\n")## logFPKM: TRUEWGCNA reports that all data are good! I now use hierarchical clustering to detect any obvious outliers. I did not see any particularly egregious outliers.
sampleclustering(log.tdata.FPKM.sample.info.subset.prefrontal.cortex.missing)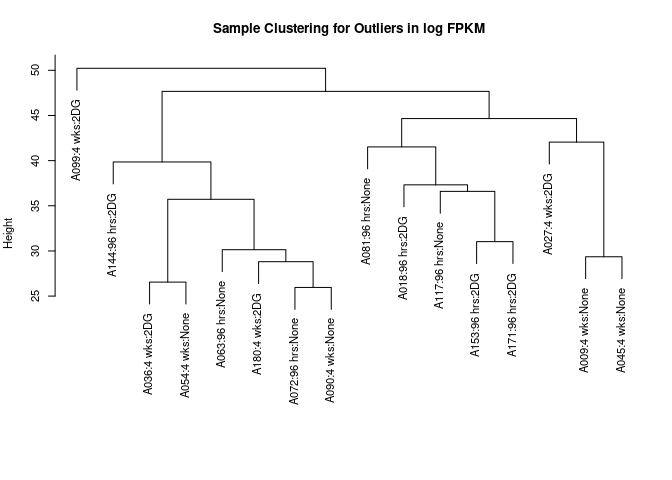
saveRDS(log.tdata.FPKM.sample.info.subset.prefrontal.cortex.missing, here("Data","Prefrontal Cortex","log.tdata.FPKM.sample.info.subset.prefrontal.cortex.missing.WGCNA.RData"))Analysis performed by Ann Wells
The Carter Lab The Jackson Laboratory 2023
ann.wells@jax.org