Summary Skeletal Muscle
Ann Wells
April 23, 2023
Summary
needed.packages <- c("tidyverse", "here", "functional", "gplots", "dplyr", "GeneOverlap", "R.utils", "reshape2","magrittr","data.table", "RColorBrewer","preprocessCore", "ARTool","emmeans", "phia", "gProfileR","rlist", "pheatmap", "WGCNA")
for(i in 1:length(needed.packages)){library(needed.packages[i], character.only = TRUE)}
source(here("source_files","WGCNA_source.R"))There were 15 samples (4 per time and treatment with one 96hr control missing due to low RNA quality). Mice were fed 2DG (6g/L) or nothing in their water for 96hrs or 4 weeks. WGCNA analysis was performed to determine gene clustering. A soft-thresholding power was chosen so that the scale-free topology correlation attempts to hit 0.9. The power chosen for skeletal muscle was 13, which allowed the correlations to hover around 0.88. This power clustered the genes into 98 modules originally, however, modules that were highly correlated (0.75 or higher) were further merged together to create 40 modules.
modules<-read.csv(here("Data","Skeletal Muscle","log.tdata.FPKM.sample.info.subset.skeletal.muscle.WGCNA.module.membership.csv"), header=T)
module_barplot(modules)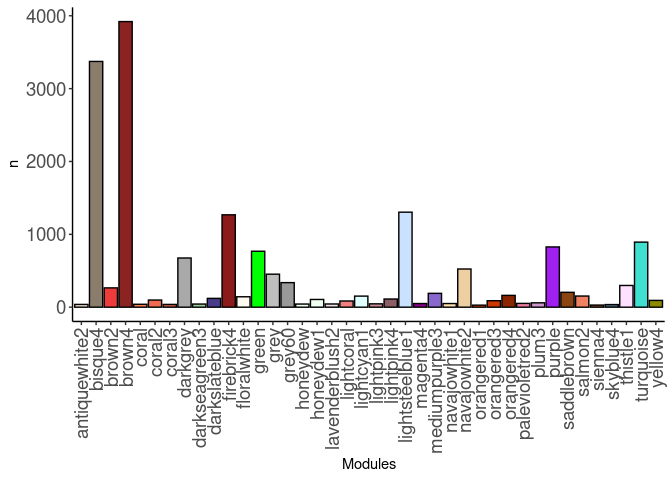
Pathways
Pathways were assessed for each module. Out of 40 modules, only 24 modules had pathway hits of any kind. The pathways that had the highest frequency across modules were metabolism of RNA, mitotic cell cycle, protein metabolism, and mRNA pathways. The brown4, darkgrey, navajowhite2, plum3, purple, salmon2, sienna4, skyblue4, and thistle1 modules all contained these pathways.
Antiquewhite2
2 pathways were identified in the Antiquewhite2 module. The pathways were cell-cell communication and nephrin family interactions.
Bisque4
86 pathways were identified in the Bisque4 module. The pathways were related to cardiac function, collagen, calcium signaling, glycogen, glycolysis, gluconeogenesis, extracellular matrix, HIF-1, JAK-STAT, NOD-like receptor, NF-kappa B, O-glycosylation, platelets, osteoclast differentiation, autoimmune disease, Th17 differentiation, TNF signaling, and toll-like receptors.
Brown2
12 Pathways were identified in the Brown2 module. The pathways were related to MAPK, metals, viruses, estrogen signaling, and heat shock.
Brown4
229 pathways were identified in the Brown 4 module. The pathways were related to autophagy, cardiac function, AMPK signaling, B cell receptor signaling, cell cycle, cellular response, chromatin, immunity, cancer, ERK, estrogen, insulin, GnRH, interleukin-17, MAPK, metabolism of RNA and proteins, mRNA, mTOR, myD88, notch signaling, osteoclast differentiation, oxidative phosphorylation, protein processing of the ER, RNA, signaling, SUMOylation, T cell receptor signaling, Th1 and 2 differentiation, Th17 differentiation, thermogenesis, TNF signaling, toll like receptors, type II diabetes, VEGF and vasopressin, ubiquitin, and WNT signaling.
Coral2
7 pathways were identified in the Coral2 module. The pathways identified were related to cardiomyopathy, muscle contraction, and cAMP signaling.
Coral3
1 pathway, long-term potentiation, was identified in the Coral3 module.
Darkgrey
82 pathways were identified in the Darkgrey module. The pathways were related to APC, cell cycle, MHC I mediated antigen processing and presentation, insulin, homologous directed recombination, metabolism, mismatch repair, p53, RUNX, ubiquitin, and TCR signaling.
Darkseagreen3
9 pathways were identified in the Darkseagreen3 module. The pathways identified were related to digestion, carbohydrate metabolism, and pancreatic and salivary secretion.
Firebrick4
3 pathways were identified in the Firebrick4 module. The pathways identified were carbon metabolism, Rap1 signaling pathway, and VEGFR2 mediated vascular permeability.
Green
12 pathways were identified in the Green module. The pathways identified were related to fatty acid metabolism, propanoate metabolism, regulation of lipolysis, and amino acid metabolism.
Grey60
6 pathways were identified in the Grey60 module. The pathways identified were related to GABA synthesis and protein-protein interactions at synapses.
Honeydew1
31 pathways were identified in the Honeydew1 module. The pathways were related to collagen, extracellular matrix, keratan, MET, platelets, and signaling.
Lightcyan1
6 pathways were identified in the lightcyan1 module. The pathways were related to neuronal function at the synapse.
Lightsteelblue1
27 pathways were identified in the lightsteelblue1 module. The pathways were related to fatty acid metabolism, mitochondrial metabolism, respiratory electron transport, TCA cycle, thermogenesis, translation, and diabetic cardiomyopathy.
Magenta4
3 pathways were identified in the Magenta4 module. The pathways were TCA cycle and signaling pathways regulating pluripotency of stem cells.
Mediumpurple3
1 pathway, metabolic pathways, was identified in the Mediumpurple3 module.
Orangered4
3 pathways were identified in the Orangered4 module. The pathways were glycosphingolipid biosynthesis-lacto and neolacto series, neuronal system, and transmission across chemical synapses.
Plum3
6 pathways were identified in the Plum3 module. The pathways were related to RNA and rRNA processing.
Purple
180 pathways were identified in the Purple module. The pathways were related to antigen processing, APC, cell cycle, cellular responses, MHC I mediated antigen processing and presentation, metabolism, mRNA, nonsense mediated decay, transcriptional regulation, ubiquitin, translation, TCA cycle, RNA polymerase II, rRNA, p53, and respiratory electron transport.
Salmon2
5 pathways were identified in the Salmon2 module. The pathways were ARMS-mediated activation, metabolism of RNA, prolonged ERK activation events, regulation of ornithine decarboxylase, and TP53 regulates transcription of additional cell cycle genes whose exact role in the p53 pathways remain uncertain.
Sienna4
9 pathways were identified in the Sienna4 module. The pathways were related to O-glycosylation, post-translational modification, and metabolism of folate and proteins.
Thistle1
15 pathways were identified in the Thistle1 module. The pathways were related to mRNA and mitotic events.
Turquoise
38 pathways were identified in the Turquoise module. The pathways were related to cholesterol, NOD-like receptors, fat digestion and absorption, drug metabolism, urea cycle, vitamin digestion and absorption, and transport.
tdata.FPKM.sample.info <- readRDS(here("Data","20190406_RNAseq_B6_4wk_2DG_counts_phenotypes.RData"))
tdata.FPKM <- readRDS(here("Data","20190406_RNAseq_B6_4wk_2DG_counts_numeric.RData"))
log.tdata.FPKM <- log(tdata.FPKM + 1)
log.tdata.FPKM <- as.data.frame(log.tdata.FPKM)
log.tdata.FPKM.sample.info <- cbind(log.tdata.FPKM, tdata.FPKM.sample.info[,27238:27240])
log.tdata.FPKM.sample.info <- log.tdata.FPKM.sample.info %>% rownames_to_column() %>% filter(rowname != "A113") %>% column_to_rownames()
log.tdata.FPKM.subset <- log.tdata.FPKM[,colMeans(log.tdata.FPKM != 0) > 0.5]
log.tdata.FPKM.subset <- log.tdata.FPKM.subset %>% rownames_to_column() %>% filter(rowname != "A113") %>% column_to_rownames()
log.tdata.FPKM.sample.info.subset.skeletal.muscle <- log.tdata.FPKM.sample.info %>% rownames_to_column() %>% filter(Tissue == "Skeletal Muscle") %>% column_to_rownames()
log.tdata.FPKM.subset <- subset(log.tdata.FPKM.sample.info.subset.skeletal.muscle, select = -c(Time,Treatment,Tissue))
WGCNA.pathway <-readRDS(here("Data","Skeletal Muscle","Chang_B6_96hr_4wk_gprofiler_pathway_annotation_list_skeletal_muscle_WGCNA.RData"))
Matched<-readRDS(here("Data","Skeletal Muscle","Annotated_genes_in_skeletal_muscle_WGCNA_Chang_B6_96hr_4wk.RData"))
module.names <- Matched$X..Module.
name <- str_split(module.names,"_")
samples <-c()
for(i in 1:length(name)){
samples[[i]] <- name[[i]][2]
}
name <- str_split(samples,"\"")
name <- unlist(name)
Treatment <- unclass(as.factor(log.tdata.FPKM.sample.info.subset.skeletal.muscle[,27238]))
Time <- unclass(as.factor(log.tdata.FPKM.sample.info.subset.skeletal.muscle[,27237]))
Treat.Time <- paste0(Treatment, Time)
phenotype <- data.frame(cbind(Treatment, Time, Treat.Time))
nSamples <- nrow(log.tdata.FPKM.sample.info.subset.skeletal.muscle)
MEs0 <- read.csv(here("Data","Skeletal Muscle","log.tdata.FPKM.sample.info.subset.skeletal.muscle.WGCNA.module.eigens.csv"),header = T, row.names = 1)
name <- str_split(names(MEs0),"_")
samples <-c()
for(i in 1:length(name)){
samples[[i]] <- name[[i]][2]
}
name <- str_split(samples,"\"")
name <- unlist(name)
colnames(MEs0) <-name
MEs <- orderMEs(MEs0)
moduleTraitCor <- cor(MEs, phenotype, use = "p");
moduleTraitPvalue <- corPvalueStudent(moduleTraitCor, nSamples)Relationship between Modules and Traits
#sizeGrWindow(10,6)
# Will display correlations and their p-values
textMatrix = paste(signif(moduleTraitCor, 2), "\n(",
signif(moduleTraitPvalue, 1), ")", sep = "");
dim(textMatrix) = dim(moduleTraitCor)
# Display the correlation values within a heatmap plot
heat <- pheatmap(moduleTraitCor, main = paste("Module-trait relationships"), display_numbers = textMatrix,color=colorRampPalette(brewer.pal(n = 11, name = "Paired"))(10), cluster_rows = F, cluster_cols = F, fontsize_number = 4, angle_col = 45, number_color = "black", border_color = "white")
heat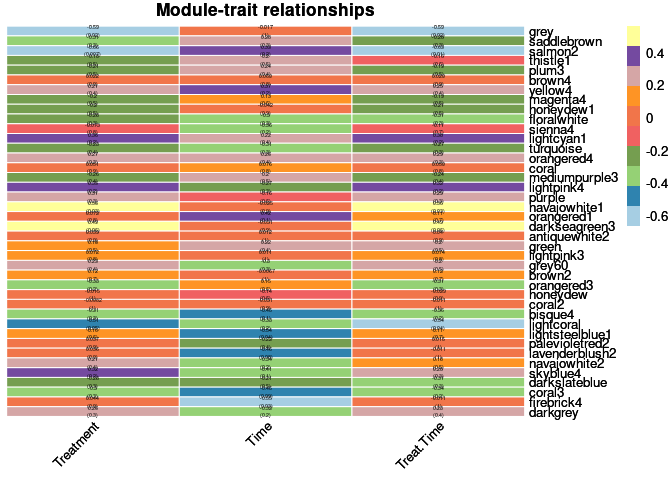
Top 10 Hub genes within each significant module
Treatment
The top 10 genes that were significant for treatment within the grey module and had the highest correlation with grey were:
- Lmbr1
- Cdk11b
- Cog3
- Rexo2
- Zfp937
- Usp46
- Rnf213
- F830208F22Rik
- Chd1
- Hspb11
When assessing the overall sample contribution through their summary eigengenes it appears that for grey most controls had a negative effect while most 2DG had a positive effect, indicating that there are many under-expressed genes for control mice and over-expressed genes 2DG treated mice.
The top 10 genes that were significant for treatment within the salmon2 module and had the highest correlation with salmon2 were:
- Nos2
- Unc5b
- Mrpl37
- Dpp9
- Pacsin2
- ENSMUSG00000097997
- Gm14326
- Hax1
- Supt7l
- Phex
When assessing the overall sample contribution through their summary eigengenes it appears that for salmon2 most controls had a negative effect while most 2DG had a positive effect, indicating that there are many under-expressed genes for control mice and over-expressed genes 2DG treated mice.
The top 10 genes that were significant for treatment within the navajowhite1 module and had the highest correlation with navajowhite1 were:
- Krt10
- Igf2bp2
- Wsb
- Etv6
- Tub
- Cbx4
- Anapc11
- Fbxo48
- 1700030K09Rik
- Mthfd1
When assessing the overall sample contribution through their summary eigengenes it appears that for navajowhite1 most controls had a positive effect while most 2DG had a negative effect, indicating that there are many over-expressed genes for control mice and under-expressed genes 2DG treated mice.
The top 10 genes that were significant for treatment within the darkseagreen3 module and had the highest correlation with darkseagreen3 were:
- Cfb
- Gm7102
- Tmem156
- Hist1h4c
- Rnf115
- Stx8
- Sap25
- Npdc1
- Erdr1
- Myo10
When assessing the overall sample contribution through their summary eigengenes it appears that for darkseagreen3 half of the controls had a positive effect while all 2DG had a negative effect, indicating that there are some over-expressed genes for control mice and under-expressed genes 2DG treated mice.
Time
The top 10 genes that were significant for time within the firebrick4 module and had the highest correlation with firebrick4 were:
- Btbd2
- Rps7
- Fam160b2
- Rnf19b
- Suv39h1
- Lama3
- Ing1
- Tmem18
- Cggbp1
- Pbxip1
When assessing the overall sample contribution through their summary eigengenes it appears that for firebrick4 most controls had a negative effect while nearly half of the 2DG mice had a positive effect, with no clear time point effects, indicating that there are some under-expressed genes for control mice and over-expressed genes 2DG treated mice.
Treatment by time
The top 10 genes that were significant for treatment by time within the grey module and had the highest correlation with grey were:
- Lmbr1
- Cog3
- Rexo2
- Zfp937
- Usp46
- Rnf213
- Llph
- F830208F22Rik
- Chd1
- Hspb11
When assessing the overall sample contribution through their summary eigengenes it appears that for grey most controls had a negative effect while most 2DG had a positive effect, independent of time point, indicating that there are many under-expressed genes for control mice and over-expressed genes 2DG treated mice.
The top 10 genes that were significant for treatment by time within the salmon2 module and had the highest correlation with salmon2 were:
- Nos2
- Gm11767
- BC003331
- Oard1
- Mrpl37
- Dpp9
- ENSMUSG00000097997
- Gm14326
- Hax1
- Dusp6
When assessing the overall sample contribution through their summary eigengenes it appears that for salmon2 most controls had a negative effect while most 2DG had a positive effect, independent of time point, indicating that there are many under-expressed genes for control mice and over-expressed genes 2DG treated mice.
The top 10 genes that were significant for treatment by time within the navajowhite1 module and had the highest correlation with navajowhite1 were:
- Krt10
- Igf2bp2
- Kbtbd12
- Wsb2
- Tub
- Cbx4
- ENSMUSG00000071047
- Anapc11
- Fbxo48
- 1700030K09Rik
When assessing the overall sample contribution through their summary eigengenes it appears that for navajowhite1 most controls had a positive effect while most 2DG had a negative effect, independent of time point, indicating that there are many over-expressed genes for control mice and under-expressed genes 2DG treated mice.
The top 10 genes that were significant for treatment by time within the darkseagreen3 module and had the highest correlation with darkseagreen3 were:
- Pank3
- Gm7102
- Tmem156
- Hist1h4c
- Rnf115
- Stx8
- Sap25
- Npdc1
- Erdr1
- Myo10
When assessing the overall sample contribution through their summary eigengenes it appears that for darkseagreen3 half of the controls had a positive effect while all 2DG had a negative effect, independent of time point, indicating that there are some over-expressed genes for control mice and under-expressed genes 2DG treated mice.
Analysis performed by Ann Wells
The Carter Lab The Jackson Laboratory 2023
ann.wells@jax.org