Contribution of Main Effects Small Intestine
Ann Wells
April 23, 2023
Introduction and Data files
This dataset contains nine tissues (heart, hippocampus, hypothalamus, kidney, liver, prefrontal cortex, skeletal muscle, small intestine, and spleen) from C57BL/6J mice that were fed 2-deoxyglucose (6g/L) through their drinking water for 96hrs or 4wks. 96hr mice were given their 2DG treatment 2 weeks after the other cohort started the 4 week treatment. The organs from the mice were harvested and processed for metabolomics and transcriptomics. The data in this document pertains to the transcriptomics data only. The counts that were used were FPKM normalized before being log transformed. It was determined that sample A113 had low RNAseq quality and through further analyses with PCA, MA plots, and clustering was an outlier and will be removed for the rest of the analyses performed. This document will determine the contribution of each main effect or their combination to each module identified, as well as the relationship of each gene within the module and its significance.
needed.packages <- c("tidyverse", "here", "functional", "gplots", "dplyr", "GeneOverlap", "R.utils", "reshape2","magrittr","data.table", "RColorBrewer","preprocessCore", "ARTool","emmeans", "phia", "gProfileR", "WGCNA","plotly", "pheatmap","ppcor", "pander","downloadthis")
for(i in 1:length(needed.packages)){library(needed.packages[i], character.only = TRUE)}
source(here("source_files","WGCNA_source.R"))
source(here("source_files","WGCNA_contribution_source.R"))tdata.FPKM.sample.info <- readRDS(here("Data","20190406_RNAseq_B6_4wk_2DG_counts_phenotypes.RData"))
tdata.FPKM <- readRDS(here("Data","20190406_RNAseq_B6_4wk_2DG_counts_numeric.RData"))
log.tdata.FPKM <- log(tdata.FPKM + 1)
log.tdata.FPKM <- as.data.frame(log.tdata.FPKM)
log.tdata.FPKM.sample.info <- cbind(log.tdata.FPKM, tdata.FPKM.sample.info[,27238:27240])
log.tdata.FPKM.sample.info <- log.tdata.FPKM.sample.info %>% rownames_to_column() %>% filter(rowname != "A113") %>% column_to_rownames()
log.tdata.FPKM.subset <- log.tdata.FPKM[,colMeans(log.tdata.FPKM != 0) > 0.5]
log.tdata.FPKM.subset <- log.tdata.FPKM.subset %>% rownames_to_column() %>% filter(rowname != "A113") %>% column_to_rownames()
log.tdata.FPKM.sample.info.subset.small.intestine <- log.tdata.FPKM.sample.info %>% rownames_to_column() %>% filter(Tissue == "Small Intestine") %>% column_to_rownames()
log.tdata.FPKM.subset <- subset(log.tdata.FPKM.sample.info.subset.small.intestine, select = -c(Time,Treatment,Tissue))
WGCNA.pathway <-readRDS(here("Data","Small Intestine","Chang_B6_96hr_4wk_gprofiler_pathway_annotation_list_small_intestine_WGCNA.RData"))
Matched<-readRDS(here("Data","Small Intestine","Annotated_genes_in_small_intestine_WGCNA_Chang_B6_96hr_4wk.RData"))
module.names <- Matched$X..Module.
name <- str_split(module.names,"_")
samples <-c()
for(i in 1:length(name)){
samples[[i]] <- name[[i]][2]
}
name <- str_split(samples,"\"")
name <- unlist(name)
Treatment <- unclass(as.factor(log.tdata.FPKM.sample.info.subset.small.intestine[,27238]))
Time <- unclass(as.factor(log.tdata.FPKM.sample.info.subset.small.intestine[,27237]))
Treat.Time <- paste0(Treatment, Time)
phenotype <- data.frame(cbind(Treatment, Time, Treat.Time))
nSamples <- nrow(log.tdata.FPKM.sample.info.subset.small.intestine)
MEs0 <- read.csv(here("Data","Small Intestine","log.tdata.FPKM.sample.info.subset.small.intestine.WGCNA.module.eigens.csv"),header = T, row.names = 1)
name <- str_split(names(MEs0),"_")
samples <-c()
for(i in 1:length(name)){
samples[[i]] <- name[[i]][2]
}
name <- str_split(samples,"\"")
name <- unlist(name)
colnames(MEs0) <-name
MEs <- orderMEs(MEs0)
moduleTraitCor <- cor(MEs, phenotype, use = "p");
moduleTraitPvalue <- corPvalueStudent(moduleTraitCor, nSamples)Relationship between Modules and Traits
#sizeGrWindow(10,6)
# Will display correlations and their p-values
textMatrix = paste(signif(moduleTraitCor, 2), "\n(",
signif(moduleTraitPvalue, 1), ")", sep = "");
dim(textMatrix) = dim(moduleTraitCor)
# Display the correlation values within a heatmap plot
heat <- pheatmap(moduleTraitCor, main = paste("Module-trait relationships"), color=colorRampPalette(brewer.pal(n = 12, name = "Paired"))(10), cluster_rows = F, cluster_cols = F, fontsize_number = 4, angle_col = 45, number_color = "black", border_color = "white")
heat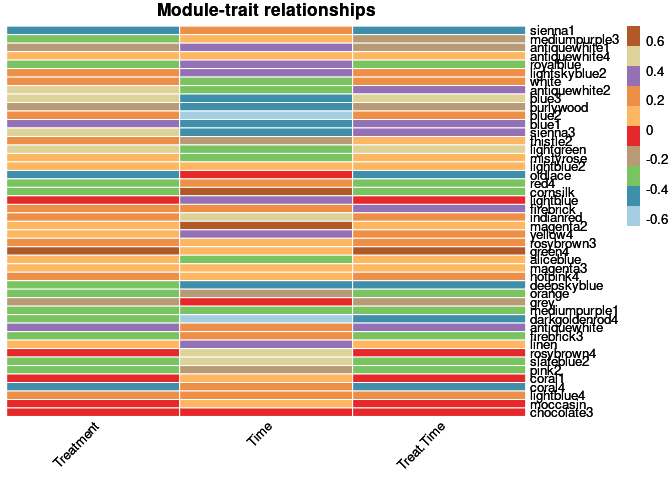
DT::datatable(moduleTraitPvalue, extensions = 'Buttons',
rownames = TRUE,
filter="top",
options = list(dom = 'Blfrtip',
buttons = c('copy', 'csv', 'excel'),
lengthMenu = list(c(10,25,50,-1),
c(10,25,50,"All")),
scrollX= TRUE), class = "display")Partial Correlation
phenotype$Treatment <- as.numeric(phenotype$Treatment)
phenotype$Time <- as.numeric(phenotype$Time)
phenotype$Treat.Time <- as.numeric(phenotype$Treat.Time)
Sigmodules <- as.data.frame(cbind(MEs$green4,MEs$blue2,MEs$cornsilk,MEs$magenta2,MEs$darkgoldenrod4))
colnames(Sigmodules) <- c("green4","blue2","cornsilk","magenta2","darkgoldenrod4")
for(i in 1:length(Sigmodules)){
cat("\n##",colnames(Sigmodules[i]),"{.tabset .tabset-fade .tabset-pills}","\n")
for(j in 1:length(phenotype)){
cat("\n###", colnames(phenotype[j]),"{.tabset .tabset-fade .tabset-pills}","\n")
for(k in 1:length(phenotype)){
cat("\n####", "Partial Correlation", colnames(phenotype[k]),"\n")
partial <- pcor.test(Sigmodules[,i], phenotype[,j],phenotype[,k])
panderOptions('knitr.auto.asis', FALSE)
print(pander(partial))
cat("\n \n")
}
cat("\n \n")
}
cat("\n \n")
}green4
Treatment
Partial Correlation Treatment
| estimate | p.value | statistic | n | gp | Method |
|---|---|---|---|---|---|
| 0.701 | 0.0036 | 3.544 | 16 | 1 | pearson |
NULL
Partial Correlation Time
| estimate | p.value | statistic | n | gp | Method |
|---|---|---|---|---|---|
| 0.7018 | 0.003545 | 3.552 | 16 | 1 | pearson |
NULL
Partial Correlation Treat.Time
| estimate | p.value | statistic | n | gp | Method |
|---|---|---|---|---|---|
| 0.03103 | 0.9126 | 0.1119 | 16 | 1 | pearson |
NULL
Time
Partial Correlation Treatment
| estimate | p.value | statistic | n | gp | Method |
|---|---|---|---|---|---|
| 0.06716 | 0.812 | 0.2427 | 16 | 1 | pearson |
NULL
Partial Correlation Time
| estimate | p.value | statistic | n | gp | Method |
|---|---|---|---|---|---|
| 0.0479 | 0.8654 | 0.1729 | 16 | 1 | pearson |
NULL
Partial Correlation Treat.Time
| estimate | p.value | statistic | n | gp | Method |
|---|---|---|---|---|---|
| -0.03103 | 0.9126 | -0.1119 | 16 | 1 | pearson |
NULL
Treat.Time
Partial Correlation Treatment
| estimate | p.value | statistic | n | gp | Method |
|---|---|---|---|---|---|
| 0.06716 | 0.812 | 0.2427 | 16 | 1 | pearson |
NULL
Partial Correlation Time
| estimate | p.value | statistic | n | gp | Method |
|---|---|---|---|---|---|
| 0.7018 | 0.003545 | 3.552 | 16 | 1 | pearson |
NULL
Partial Correlation Treat.Time
| estimate | p.value | statistic | n | gp | Method |
|---|---|---|---|---|---|
| 0.7022 | 0.003513 | 3.556 | 16 | 1 | pearson |
NULL
blue2
Treatment
Partial Correlation Treatment
| estimate | p.value | statistic | n | gp | Method |
|---|---|---|---|---|---|
| 0.2651 | 0.3396 | 0.9914 | 16 | 1 | pearson |
NULL
Partial Correlation Time
| estimate | p.value | statistic | n | gp | Method |
|---|---|---|---|---|---|
| 0.3494 | 0.2018 | 1.345 | 16 | 1 | pearson |
NULL
Partial Correlation Treat.Time
| estimate | p.value | statistic | n | gp | Method |
|---|---|---|---|---|---|
| 0.6882 | 0.004562 | 3.42 | 16 | 1 | pearson |
NULL
Time
Partial Correlation Treatment
| estimate | p.value | statistic | n | gp | Method |
|---|---|---|---|---|---|
| -0.6755 | 0.005718 | -3.303 | 16 | 1 | pearson |
NULL
Partial Correlation Time
| estimate | p.value | statistic | n | gp | Method |
|---|---|---|---|---|---|
| -0.6513 | 0.008535 | -3.095 | 16 | 1 | pearson |
NULL
Partial Correlation Treat.Time
| estimate | p.value | statistic | n | gp | Method |
|---|---|---|---|---|---|
| -0.6882 | 0.004562 | -3.42 | 16 | 1 | pearson |
NULL
Treat.Time
Partial Correlation Treatment
| estimate | p.value | statistic | n | gp | Method |
|---|---|---|---|---|---|
| -0.6755 | 0.005718 | -3.303 | 16 | 1 | pearson |
NULL
Partial Correlation Time
| estimate | p.value | statistic | n | gp | Method |
|---|---|---|---|---|---|
| 0.3494 | 0.2018 | 1.345 | 16 | 1 | pearson |
NULL
Partial Correlation Treat.Time
| estimate | p.value | statistic | n | gp | Method |
|---|---|---|---|---|---|
| 0.199 | 0.477 | 0.7322 | 16 | 1 | pearson |
NULL
cornsilk
Treatment
Partial Correlation Treatment
| estimate | p.value | statistic | n | gp | Method |
|---|---|---|---|---|---|
| -0.3696 | 0.1751 | -1.434 | 16 | 1 | pearson |
NULL
Partial Correlation Time
| estimate | p.value | statistic | n | gp | Method |
|---|---|---|---|---|---|
| -0.4499 | 0.09245 | -1.816 | 16 | 1 | pearson |
NULL
Partial Correlation Treat.Time
| estimate | p.value | statistic | n | gp | Method |
|---|---|---|---|---|---|
| -0.6356 | 0.01088 | -2.968 | 16 | 1 | pearson |
NULL
Time
Partial Correlation Treatment
| estimate | p.value | statistic | n | gp | Method |
|---|---|---|---|---|---|
| 0.6136 | 0.01499 | 2.802 | 16 | 1 | pearson |
NULL
Partial Correlation Time
| estimate | p.value | statistic | n | gp | Method |
|---|---|---|---|---|---|
| 0.5701 | 0.02649 | 2.502 | 16 | 1 | pearson |
NULL
Partial Correlation Treat.Time
| estimate | p.value | statistic | n | gp | Method |
|---|---|---|---|---|---|
| 0.6356 | 0.01088 | 2.968 | 16 | 1 | pearson |
NULL
Treat.Time
Partial Correlation Treatment
| estimate | p.value | statistic | n | gp | Method |
|---|---|---|---|---|---|
| 0.6136 | 0.01499 | 2.802 | 16 | 1 | pearson |
NULL
Partial Correlation Time
| estimate | p.value | statistic | n | gp | Method |
|---|---|---|---|---|---|
| -0.4499 | 0.09245 | -1.816 | 16 | 1 | pearson |
NULL
Partial Correlation Treat.Time
| estimate | p.value | statistic | n | gp | Method |
|---|---|---|---|---|---|
| -0.3111 | 0.2591 | -1.18 | 16 | 1 | pearson |
NULL
magenta2
Treatment
Partial Correlation Treatment
| estimate | p.value | statistic | n | gp | Method |
|---|---|---|---|---|---|
| 0.04077 | 0.8853 | 0.1471 | 16 | 1 | pearson |
NULL
Partial Correlation Time
| estimate | p.value | statistic | n | gp | Method |
|---|---|---|---|---|---|
| 0.05394 | 0.8486 | 0.1948 | 16 | 1 | pearson |
NULL
Partial Correlation Treat.Time
| estimate | p.value | statistic | n | gp | Method |
|---|---|---|---|---|---|
| -0.6511 | 0.008561 | -3.093 | 16 | 1 | pearson |
NULL
Time
Partial Correlation Treatment
| estimate | p.value | statistic | n | gp | Method |
|---|---|---|---|---|---|
| 0.6553 | 0.008005 | 3.128 | 16 | 1 | pearson |
NULL
Partial Correlation Time
| estimate | p.value | statistic | n | gp | Method |
|---|---|---|---|---|---|
| 0.6548 | 0.008076 | 3.123 | 16 | 1 | pearson |
NULL
Partial Correlation Treat.Time
| estimate | p.value | statistic | n | gp | Method |
|---|---|---|---|---|---|
| 0.6511 | 0.008561 | 3.093 | 16 | 1 | pearson |
NULL
Treat.Time
Partial Correlation Treatment
| estimate | p.value | statistic | n | gp | Method |
|---|---|---|---|---|---|
| 0.6553 | 0.008005 | 3.128 | 16 | 1 | pearson |
NULL
Partial Correlation Time
| estimate | p.value | statistic | n | gp | Method |
|---|---|---|---|---|---|
| 0.05394 | 0.8486 | 0.1948 | 16 | 1 | pearson |
NULL
Partial Correlation Treat.Time
| estimate | p.value | statistic | n | gp | Method |
|---|---|---|---|---|---|
| 0.1057 | 0.7077 | 0.3833 | 16 | 1 | pearson |
NULL
darkgoldenrod4
Treatment
Partial Correlation Treatment
| estimate | p.value | statistic | n | gp | Method |
|---|---|---|---|---|---|
| -0.3646 | 0.1815 | -1.412 | 16 | 1 | pearson |
NULL
Partial Correlation Time
| estimate | p.value | statistic | n | gp | Method |
|---|---|---|---|---|---|
| -0.446 | 0.09562 | -1.797 | 16 | 1 | pearson |
NULL
Partial Correlation Treat.Time
| estimate | p.value | statistic | n | gp | Method |
|---|---|---|---|---|---|
| 0.5917 | 0.02016 | 2.646 | 16 | 1 | pearson |
NULL
Time
Partial Correlation Treatment
| estimate | p.value | statistic | n | gp | Method |
|---|---|---|---|---|---|
| -0.6186 | 0.01395 | -2.839 | 16 | 1 | pearson |
NULL
Partial Correlation Time
| estimate | p.value | statistic | n | gp | Method |
|---|---|---|---|---|---|
| -0.5761 | 0.02461 | -2.541 | 16 | 1 | pearson |
NULL
Partial Correlation Treat.Time
| estimate | p.value | statistic | n | gp | Method |
|---|---|---|---|---|---|
| -0.5917 | 0.02016 | -2.646 | 16 | 1 | pearson |
NULL
Treat.Time
Partial Correlation Treatment
| estimate | p.value | statistic | n | gp | Method |
|---|---|---|---|---|---|
| -0.6186 | 0.01395 | -2.839 | 16 | 1 | pearson |
NULL
Partial Correlation Time
| estimate | p.value | statistic | n | gp | Method |
|---|---|---|---|---|---|
| -0.446 | 0.09562 | -1.797 | 16 | 1 | pearson |
NULL
Partial Correlation Treat.Time
| estimate | p.value | statistic | n | gp | Method |
|---|---|---|---|---|---|
| -0.4201 | 0.119 | -1.669 | 16 | 1 | pearson |
NULL
Scatterplots Module membership vs. Gene significance
Time
time.contribution(phenotype, log.tdata.FPKM.subset, MEs)sienna1
mediumpurple3
antiquewhite1
antiquewhite4
royalblue
lightskyblue2
white
antiquewhite2
blue3
burlywood
blue2
blue1
sienna3
thistle2
lightgreen
mistyrose
lightblue2
oldlace
red4
cornsilk
lightblue
firebrick
indianred
magenta2
yellow4
rosybrown3
green4
aliceblue
magenta3
hotpink4
deepskyblue
orange
grey
mediumpurple1
darkgoldenrod4
antiquewhite
firebrick3
linen
rosybrown4
slateblue2
pink2
coral1
coral4
lightblue4
moccasin
chocolate3
Treatment
treat.contribution(phenotype, log.tdata.FPKM.subset, MEs)sienna1
mediumpurple3
antiquewhite1
antiquewhite4
royalblue
lightskyblue2
white
antiquewhite2
blue3
burlywood
blue2
blue1
sienna3
thistle2
lightgreen
mistyrose
lightblue2
oldlace
red4
cornsilk
lightblue
firebrick
indianred
magenta2
yellow4
rosybrown3
green4
aliceblue
magenta3
hotpink4
deepskyblue
orange
grey
mediumpurple1
darkgoldenrod4
antiquewhite
firebrick3
linen
rosybrown4
slateblue2
pink2
coral1
coral4
lightblue4
moccasin
chocolate3
Treatment by Time
treat.time.contribution(phenotype, log.tdata.FPKM.subset, MEs)sienna1
mediumpurple3
antiquewhite1
antiquewhite4
royalblue
lightskyblue2
white
antiquewhite2
blue3
burlywood
blue2
blue1
sienna3
thistle2
lightgreen
mistyrose
lightblue2
oldlace
red4
cornsilk
lightblue
firebrick
indianred
magenta2
yellow4
rosybrown3
green4
aliceblue
magenta3
hotpink4
deepskyblue
orange
grey
mediumpurple1
darkgoldenrod4
antiquewhite
firebrick3
linen
rosybrown4
slateblue2
pink2
coral1
coral4
lightblue4
moccasin
chocolate3
Analysis performed by Ann Wells
The Carter Lab The Jackson Laboratory 2023
ann.wells@jax.org